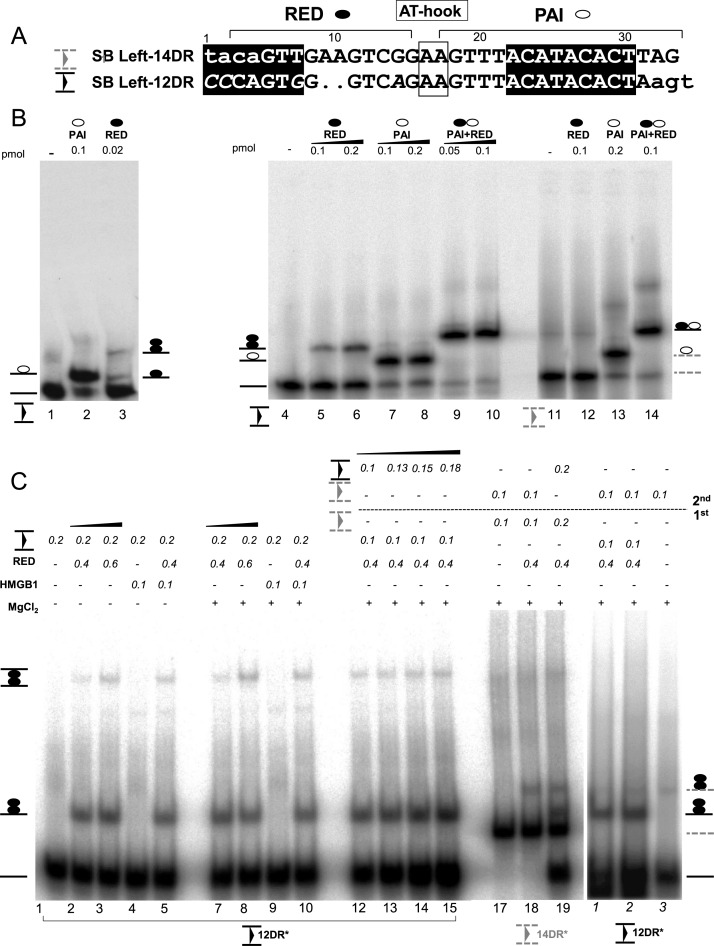
Figure 3.
Modeling second-end-capture (SEC) of Sleeping Beauty in gel mobility shift assay (EMSA). (A) Alignment of the 14DR (outer, filled triangle between two lines) and 12DR (inner) of the left inverted repeat (IR). The nucleotides involved in DNA-protein interaction (21,25), are shown in uppercase, while the nonidentical nucleotides are in italics. The nucleotides recognized by PAI (empty circle) or RED (black circle) subdomains, and the AT-hook (framed) are indicated (21). The nucleotides resembling the ‘heptamer’ and ‘nonamer’ motifs of the RAG1 (50) are highlighted in black boxes. The length of the spacer between motifs is 12 bp, or 14 bp in the inner and outer DR, respectively. The 2 bp differences between inner and outer DRs are dotted. (B) DNA binding properties of RED (N58-123, black circle), PAI (N157, empty circle) or the full N-terminal DNA binding domain (PAI+RED) were characterized by EMSA. Upper panels: labeled oligonucleotides corresponding to the 12DR (black triangle, plain lines), the 14DR (gray triangle, dashed lines) or the 12+AA DR (dotted, black) were used as DNA substrates. The schematic of the predicted nucleoprotein complexes are depicted. Uncomplexed DNA—plain line. Complexes formed with the full N-terminal DNA binding domain (N123, PAIRED) were used as size markers (∼2×RED). The complexes were separated on 4% native gels. Note that monomeric RED–12DR complexes can be detected only at low protein concentration (0.02 versus 0.1 pmol, left panel, lane 3). (C) The 12DR/12DR rule (a selective capture of 12DR by the 12DR–RED complex) of Sleeping Beauty second-end-capture (SEC). HMGB1 promotes RED–12DR formation in a Mg2+ independent manner (lanes 1–10). Nucleoprotein complexes of 12DR–RED formed on 12DR (0.2 pmol) at various concentrations of RED indicated in the absence or presence of HMGB1 (μM). See also Supplementary Figure S3C. Asterisks indicate radioactively labeled substrate. Note that the formation of the low mobility band does not require the presence of MgCl2 (5 mM). Investigating SEC by staged EMSA (first–second). (First) 12DR substrate was pre-incubated with RED, (second) followed by the addition of labeled 12DR (lanes 12–15), or 14DR partner (right panel, 1–3), respectively (15 min, followed by 10 min on ice). Binding reactions were performed at constant RED concentration (0.4 pmol). Note that detectable SEC formation was neither detected when 14DR was offered as a partner to 12DR (right panel, 1–3), or in a reciprocal experiment, when complex formation was initiated on 14DR (lanes 17–19). The complexes were separated on 4% native gels.