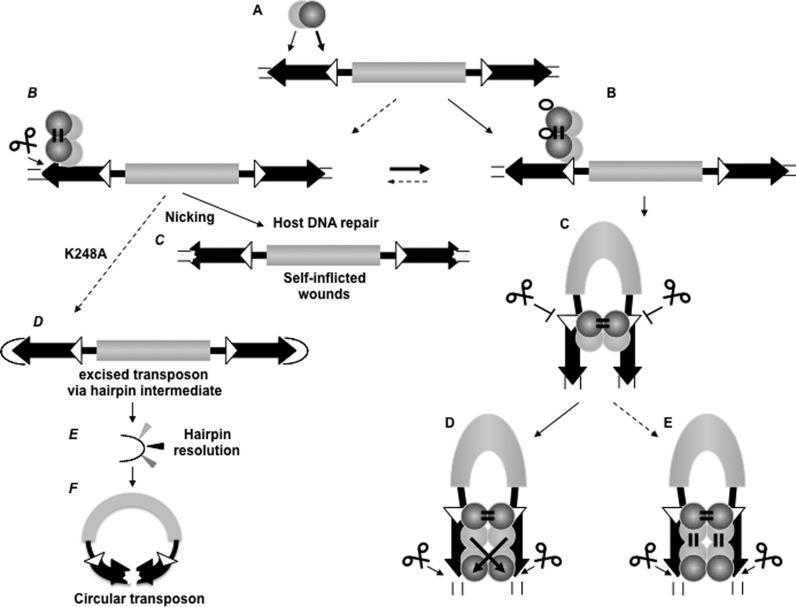
Figure 6.
The ordered assembly of Sleeping Beauty transposition enforces paired-end complex formation (model). (A) Primary contact, substrate recognition by the transposase. Left IR is preferred over the right one. The transposase is able to recognize both the inner and outer DRs primarily by PAI (light gray ball), while RED (dark gray ball) is involved in the primary substrate identification. (B) RED is able to differentiate between 12DR (white triangle) versus 14DR (black triangle), and transposase binding is guided to 12DR. In the presence of HMGB1 (empty ellipse), the preference for 12DR-binding is further emphasized. The transposase dimerizes rapidly upon binding to 12DR (or binds as a dimer) via RED homo-dimerization. (C) The dimerized transposase captures the right IR in a naked form, governed by the 12/12DR rule. The second end capture complex (SEC) involves the 12DRs, and cleavage is strictly inhibited at this stage of the reaction. SEC formation is promoted by the presence of HMGB1, but Mg2+ is not required (not shown). (D and E) 14DRs are positioned in the complex for cleavage, presumably involving PAI-homo-dimerization and 14DR–RED interaction. The configuration of the complex can be trans-crossed (D) or trans-parallel (E) (43). PAI-enhancer interaction (21) is predicted to stabilize the pre-integration complex (not shown). Note: Since both PAI and RED are involved in dual function, conformational changes might coordinate the DNA-binding and protein dimerization activities (not shown). Left panel: (B) Dotted arrow indicates that the under suboptimal condition and/or by transposase variants (hexagons) generated by genetic drift, nicking occurs at 14DRs (instead of double strand cleavage). Nicking is also preceded by paired end formation, and might liberate the transposon (as in F). (C) The DNA repair machinery of the host diversifies the DNA sequence around the nick (‘self-inflicted wounds’). (D) K248A liberates the transposons via hairpin resolution. (E) Hairpin resolution diversifies the IRs. (F) Hairpin resolution seals the transposon to form an extrachromosomal circle. Circular transposon molecules have markedly reduced integration ability, but might be ideal for horizontal transfer. Note: For simplicity, the catalytic domain of the transposase and the enhancer motif are not shown, and HMGB1 is shown only in B. Possible conformational changes between steps are not shown.