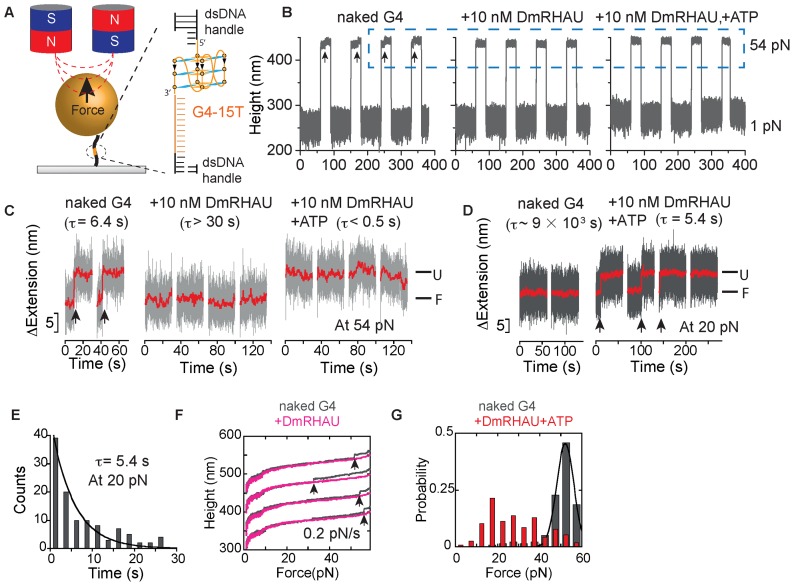
Figure 1.
(A) Schematic of the magnetic tweezers setup and the G4-15T DNA construct. A Myc2345 ssDNA sequence (G4-15T, orange) is flanked by two dsDNA handles (489 bp and 601 bp, black) and is tethered between a paramagnetic bead and a coverslip. The magnified area shows the local region containing the G4-15T sequence folded into a G4 structure. (B) Representative time trace of the bead height change during force jump cycles between 1 pN and 54 pN measured without DmRHAU, with 10 nM DmRHAU but without ATP, and with both 10 nM DmRHAU and 1 mM ATP, as indicated in the figure panel. (C) Representative time trace of extension change of a G4 molecule (grey data) after jumping to 54 pN in several force-jump cycles taken from data in Figure 1B. The extensions corresponding to folded (F) and unfolded (U) G4 are also indicated; G4 unfolding events are indicated by arrows. (D) Representative time trace of extension change of a G4 molecule after jumping to 20 pN in several force-jump cycles of 1 pN (60 s) to 20 pN (60 s) to 54 pN (30 s), in the absence (left) and presence (right) of DmRHAU and ATP. Unfolding events are indicated by arrows. In (C and D), red traces are smoothed data using adjacent averages with a 0.5-s sliding window. (E) Histogram of the lifetime of G4-15T at 20 pN (110 data obtained from 10 independent molecules) in the presence of 10 nM DmRHAU and 1 mM ATP. (F) Force-extension curve of G4-15T in the absence (grey) and presence (pink) of DmRHAU. (G) Unfolding force distribution of G4-15T in the absence (grey) and presence (red) of DmRHAU and ATP.