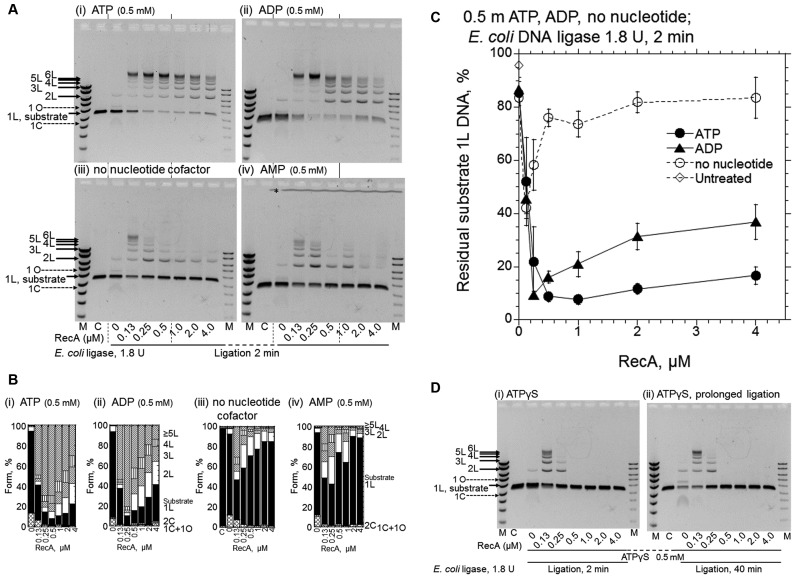
Figure 4.
ADP is also an effective cofactor for RecA-enhanced DNA ligation by E. coli DNA ligase, but ATPγS causes the suppression of the DNA ligation. (A) Representative gel-profiles of the electrophoretic analysis of DNA products formed. (B) Fractions of the substrates and products quantified from the gel profiles in A shown by cumulative bar graphs. After the preincubation of the linear dsDNA with a 3′ four-nucleotide overhang with the indicated amounts of RecA and nucleotide cofactors for 5 min, the ligation was initiated by the addition of E. coli DNA ligase (1.8 U) without temperature shifts followed by a 2 min incubation. Cofactors added: panel (i), ATP; panel (ii), ADP; panel (iii), without nucleotide cofactor; panel (iv), AMP; Lanes C, untreated linear substrate dsDNA. Lanes M, DNA size and amount markers are the same as in Figure 1B. * in panel (iv) in A, see Figure 2. Symbols of the forms of the products and substrate in B are indicated in Figure 1B. (C) Quantified representation of the fractions of residual substrate dsDNA (1L) against the amounts of RecA. Each symbol represents the average and standard deviation of the results from at least three independent experiments, including those shown in the gel profiles (i–iii) shown in A. • (closed circles), with ATP; ▴ (closed triangles), with ADP; ∘ (open circles), no nucleotide cofactors; ◊ (open diamonds), untreated substrate linear dsDNA (1L) quantified from gel profile (iii), lane labeled C. (D) Effects of ATPγS. The conditions of the reactions were the same as in A, with ATPγS. The periods of the DNA ligation-reaction were 2 min in panel (i) and 40 min in panel (ii). We confirmed the reproducibility of the effects of AMP in A and ATPγS in D by repeating the experiments twice and four times, respectively.