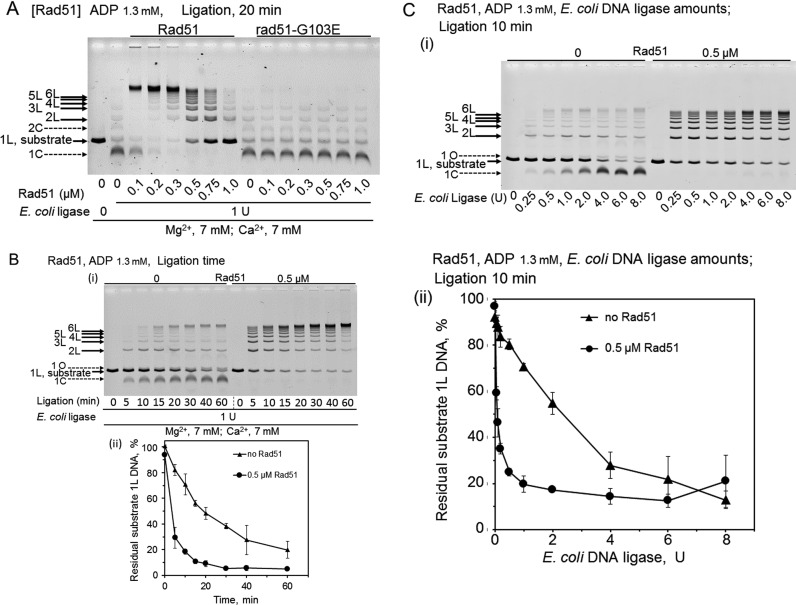
Figure 7.
Features of ADP-dependent Rad51-enhanced DNA ligation and the effects of the G103E-substitution in Rad51. CaCl2 (7 mM) were added to the standard reaction mixtures (including 7 mM MgCl2). Linear dsDNA with a 3′ four-nucleotide overhang was incubated with Rad51 (wild-type) or rad51-G103E for 5 min, before the addition of E. coli DNA ligase to start the DNA ligation without temperature shifts, followed by an incubation for the indicated time. (A) Effects of the amounts of Rad51 and rad51-G103E in the presence of ADP. The linear dsDNA was treated with 1 U E. coli DNA ligase for 20 min, in the presence of the indicated amounts of Rad51 or rad51-G103E and ADP. We repeated these experiments three times to confirm the reproducibility. (B) Effects of the incubation times with or without Rad51 in the presence of ADP. The linear dsDNA was treated with 1 U E. coli DNA ligase, in the presence of ADP, and 0 or 0.5 μM Rad51 for the indicated time. The graph attached is the quantified representation of the fractions of substrate dsDNA (1L). Each symbol is the average of results obtained from two independent experiments. Error bars show the smaller and larger values, and not standard deviations only in this figure. • (closed circles), with 0.5 μM Rad51; ▴ (closed triangles), without Rad51. We repeated these experiments twice to confirm the reproducibility. (C) Effects of the amounts of E. coli DNA ligase in the presence or absence of 0.5 μM Rad51. The linear dsDNA was treated with the indicated amounts of E. coli DNA ligase in the presence of ADP and Rad51 for 10 min. (i) Representative gel-profiles of the electrophoretic analysis of DNA products. (ii) Quantified representation of the fractions of residual substrate dsDNA (1L). Each symbol is the average of results obtained from at least three independent experiments. Error bars show standard deviations. • (closed circles), with 0.5 μM Rad51; ▴ (closed triangles), without Rad51.