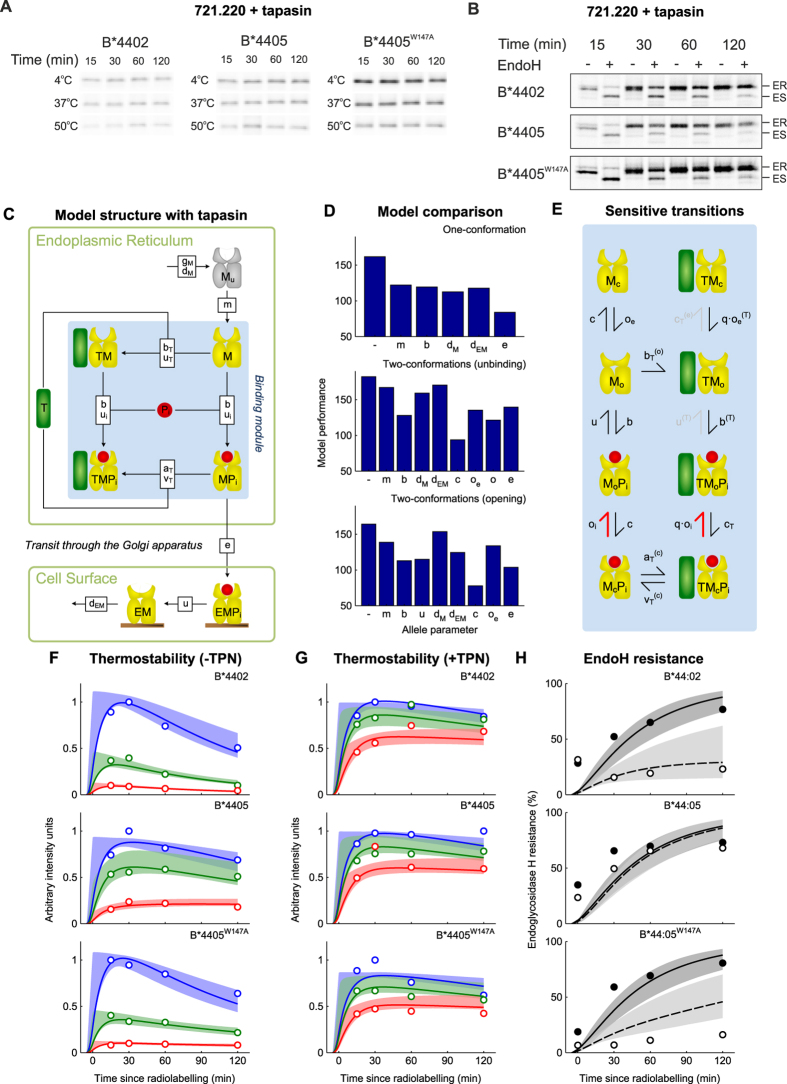
Figure 6. Computational systems models of the mechanisms of MHC I peptide selection, fit to in vivo peptide selection data for B*4402, B*4405 and B*4405W147A in the presence and absence of tapasin.
(A) Repeating the thermostability assay shown in Fig. 2A in the presence of tapasin indicates that B*4402 and B*4405W147A now acquire thermostability equal to that of B*4405 (quantified in G). (B) Repeating the pulse chase assay shown in Fig. 2B in the presence of tapasin shows that all alleles select high affinity peptides in the presence of tapasin and traffic to the cell surface (quantified in H). (C) Graphical depiction of the general computational systems model of the mechanisms of peptide selection in the presence of tapasin, as in Fig. 2D. (D) Comparison of model performance for one-conformation and two-conformation models, as in Fig. 2E. The most likely model (with the lowest BIC score) was once again identified as the two-conformation model with MHC I opening as the peptide-dependent step and MHC I closing rate c as the allele-dependent parameter. (E) Flux analysis of the two-conformation model with peptide-dependent opening (including tapasin) reveals an anti-clockwise cycle of tapasin mediated peptide editing (the peptide-specific reactions are shown as thick red symbols, and grey lines indicate unfavorable reactions). (F–H) Comparison of model behavior including a function for tapasin, analogous to Fig. 2F,G. Experimental measurements (circles) quantified from Figs 2A,B and 4A,B and panels (A,B) in this figure. In H solid black are + tapasin and dashed/open are – tapasin.