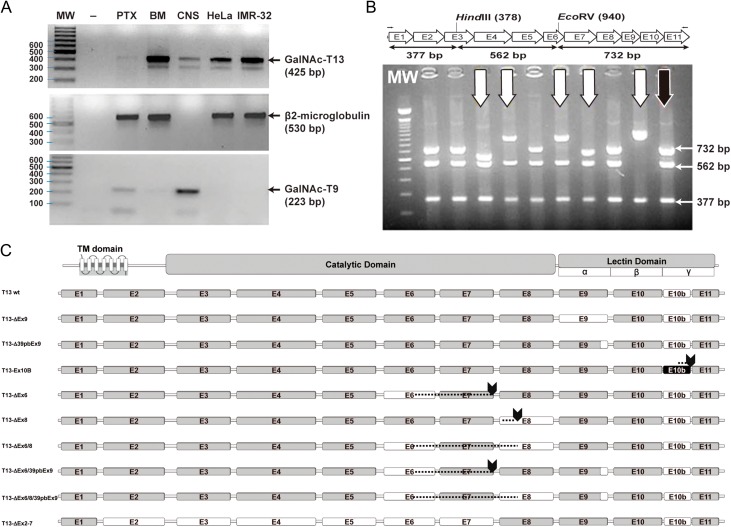
Fig. 2.
GalNAc-T13 splice variants. (A) Reverse transcription-polymerase chain reaction (RT-PCR) was performed with GalNT13_Fw and T13_XhoI_Rev primers on four different cancer cell lines and on human brain tissue. Several bands of unexpected size were observed after ethidium bromide gel staining. MW: molecular weight (100 bp DNA ladder); -: negative control (water); IGR-N-91 PTX: neuroblastoma cell line derived from a primary tumor; IGR-N-91 BM: metastatic neuroblastoma cell line; CNS: central nervous system tissue (brain); HeLa: cervical cancer cell line; IMR-32: neuroblastoma cell line. The β2M (β2-microglobulin) gene was amplified to verify cDNA quality (except for CNS, where we used GalNAc-T9 as a control). (B) Screening of GalNAc-T13 splice variants by colony PCR. Splice variants were screened by RT-PCR with T13_XhoI_Fw and T13_XhoI_Rev primers. The product was cloned into the pGEM-T Easy plasmid and analyzed by colony PCR and digestion with restriction enzymes HindIII and EcoRV. The expected product sizes were 732, 562 and 377 bp. In the illustrative gel (representative gel of 9 of more than 300 analyzed clones), the black arrow indicates digestion products of the previously characterized positive control (pGEM-T with the GalNAc-T13 coding region, confirmed by sequencing), whereas the white arrows mark clones with alternative digestion patterns. These clones were subsequently analyzed by sequencing. (C) Exon organization of the newly identified GalNAc-T13 splice variants. The transmembrane (TM), catalytic and lectin domains of GalNAc-Ts are indicated, as well as the three lectin subdomains α, β and γ. The amino acid (AA) extension of each variant is indicated on the right. Skipped exons are shown in white, Ex10b is shown in black and stop codons are marked by arrows. Dashed lines indicate regions where exon skipping introduces a change in the reading frame, resulting in a different AA sequence.