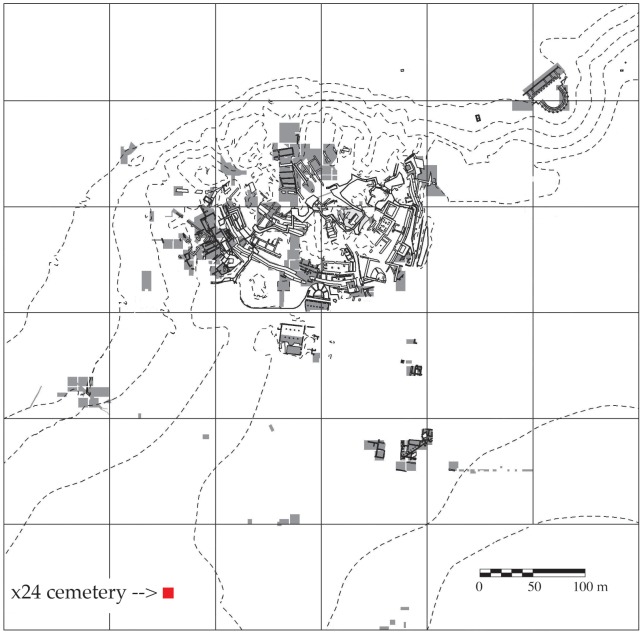
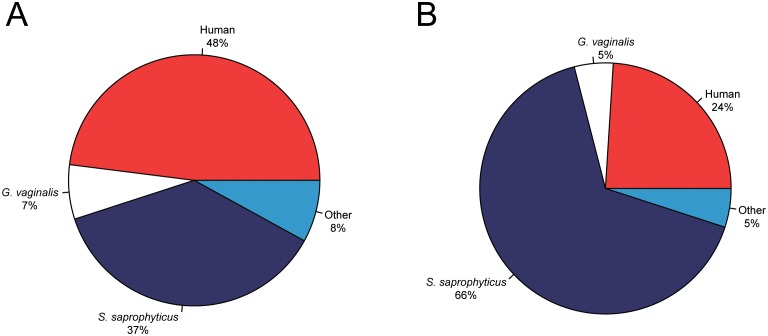
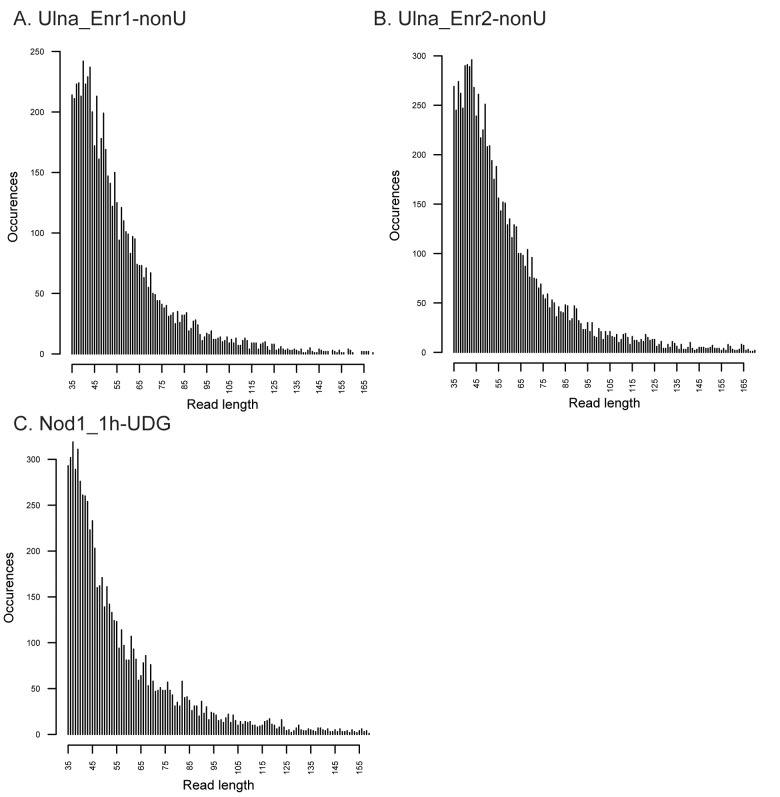
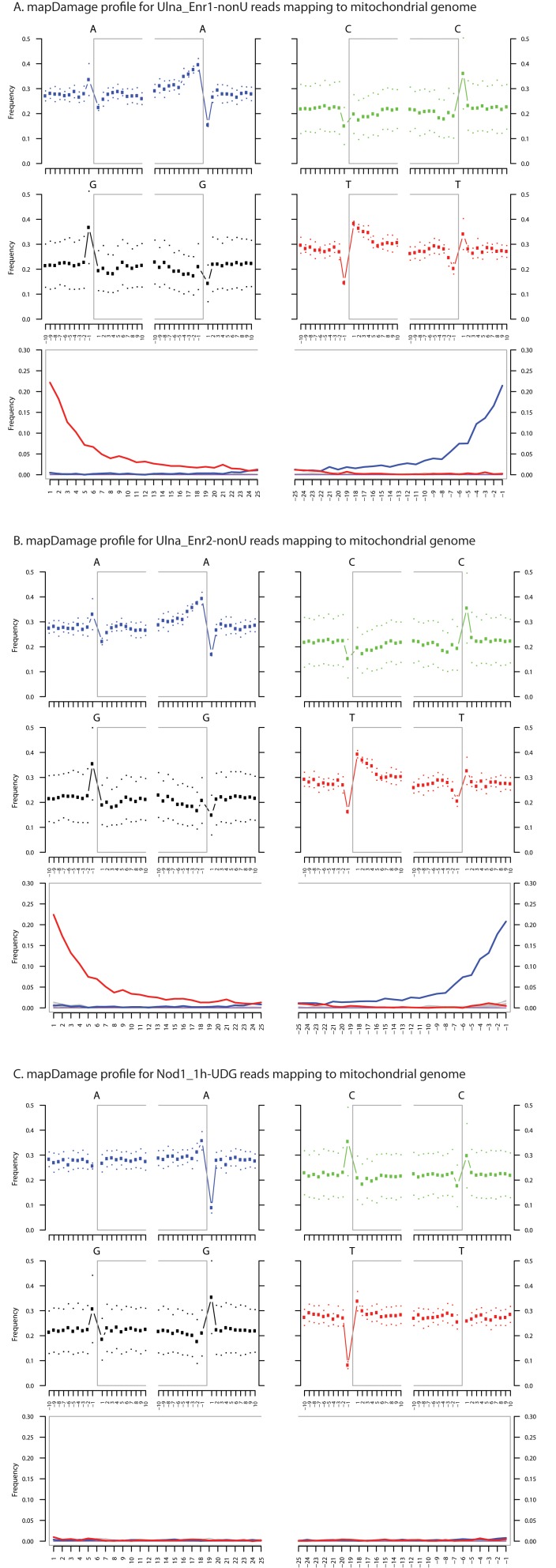
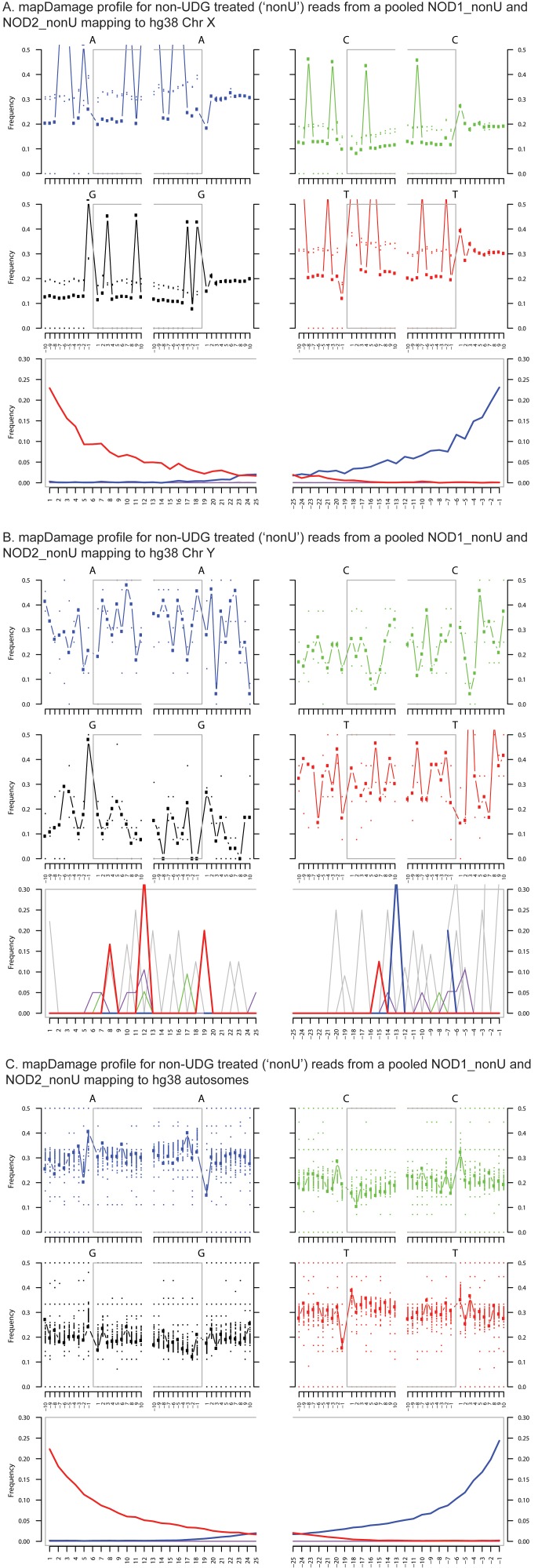
Figure 1. Calcified nodule found among the skeletal remains at Troy.
(A) Burial x24.177 (grave 14, cemetery in quadrat x24). Photo credit Gebhard Bieg, 2005. (B) Cross-section of nodule (sample no x24.177), photo credit: Pathologie Nordhessen 2009. Scale represents 1 cm. (C) Location of Troy. Modern day Turkey is shaded in gray.