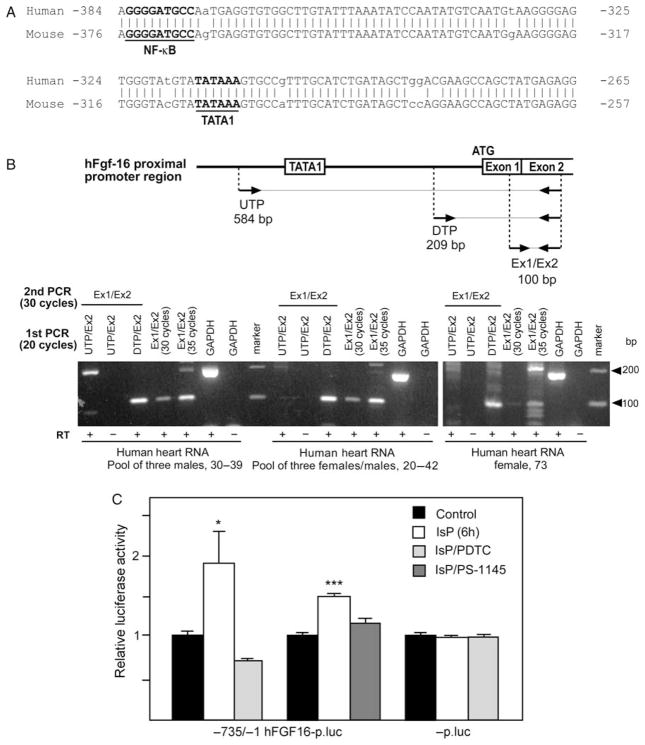
Figure 4.
(A) Alignment of human (−383/−267) and mouse (−375/−259) Fgf-16 gene sequences. Positions of putative TATA1 and NF-κB DNA sequences are shown. Mismatched sequences are indicated (no vertical bar/lower case). (B) Schematic showing the relative positions of the upstream (UTP) and downstream TATA1 primer (DTP) as well as the exon 1 and exon 2 (Ex1/Ex2) primers, together with expected amplicon sizes after RT–PCR. Three human (h) heart RNA samples were assessed by RT–PCR using Ex1/Ex2 primers for 30 and 35 cycles. Results are visualized by electrophoresis and ethidium bromide staining. To assess whether the ‘TATA’ is likely part of the hFGF-16 promoter region, RNA was amplified (RT–PCR) using UTP vs. DTP partnered with an exon 2 (Ex2) primer for 20 cycles. A second round of PCR using Ex1/Ex2 primers (30 cycles) was used to detect FGF-16 RNA as a 100 bp product. GAPDH RNA was also assessed (±RT) as a control. The mobility of a 100 bp marker is shown. (C) Neonatal rat cardiomyocytes were transfected with −735/−1 hFGF-16p.luc or -p.luc, and treated without or with IsP for 6 h, and without or with PDTC or PS-1145 pretreatment. Results are expressed relative to luciferase activity in untreated (Control) cultures, which has been arbitrarily set to 1.0. Basal levels of activity for −735/−1 hFGF-16p.luc and -p.luc are 7.28 ±0.08 and 11.42 ±0.08, respectively, and are means ±SEM (n = 3).