Figure 6. Increased activation of the Imd pathway and susceptibility to bacterial infection in miR-263a mutants.
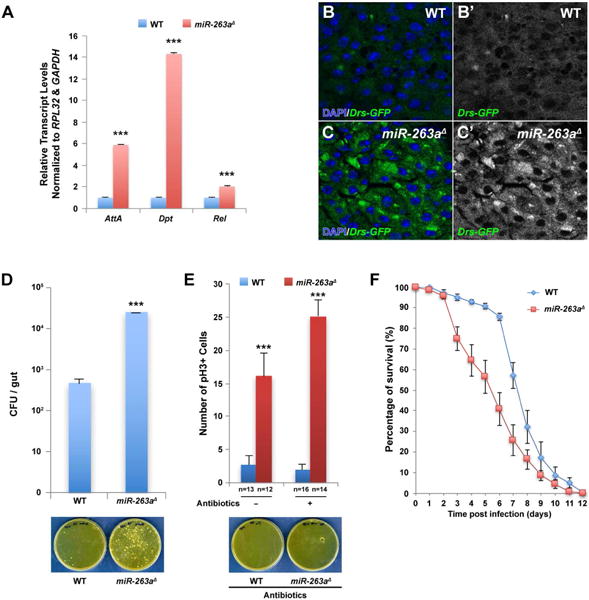
(A) Significant increase in the relative transcript levels of the AMPs was detected in miR-263a mutant midguts by qPCR. (B–C′) Expression of Drs-GFP in the posterior midgut. The expression of Drs-GFP is significantly lower in wild-type (B-B′) than in miR-263a mutants (C-C′). (D) Internal bacterial load significantly increased in the miR-263a mutants. Error bars indicate SEM. ***P < 0.001 (two-tailed t-test). Wild-type and miR-263a mutant flies homogenates were spread on the plate. The bacteria colonies were shown on the bottom panel. (E) The average number of pH3+ cells in the posterior midguts after antibiotics treatment for 16 days. Feeding antibiotics had no effect on the hyperproliferation of ISCs observed in the miR-263a mutant midguts. Bottom panel shows the bacteria colonies on medium plate spread with antibiotics fed wild-type and miR-263a mutant homogenate. Internal bacteria were absent in both wild-type and miR-263a mutant flies fed for 16 days with food containing antibiotics. “n” denotes the number of posterior midguts examined for each genotype. Error bars indicate SEM. ***P < 0.001 (two-tailed t-test). (F) Survival analysis of wild-type and miR-263a mutant flies upon oral infection with P. aeruginosa. The miR-263a mutant flies exhibited significant increase in lethality 3 days after infection. Error bars indicate SEM. See also Figure S6.
