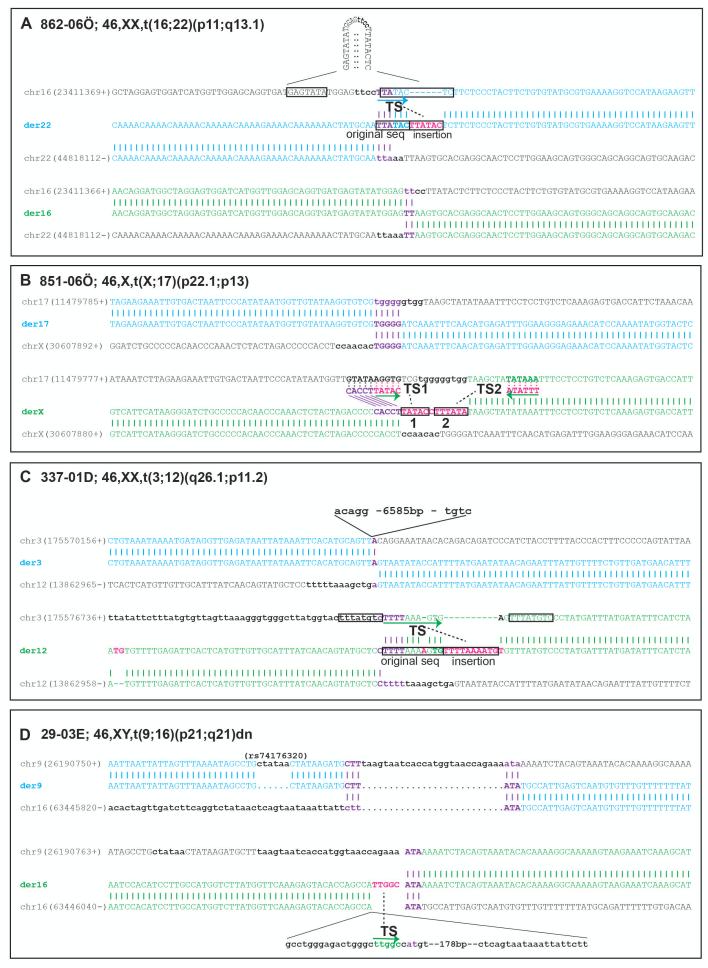
Figure 3. Four additional translocation carriers with evidence for templated insertions originating from nearby genomic segments.
Sequence alignments of the derivative chromosome sequences to the corresponding regions on the parental chromosomes. Deletions are shown in lower case bold letters. Microhomology is highlighted in purple with the most plausible parental chromosome indicated by bold text. Insertions and SNVs are shown in pink (A) Sequence alignments from case 862-06Ö. Derivative chromosome 22 (der22) is shown on top and derivative chromosome 16 (der16) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr16 and chr22) sequences are labeled in blue for der22 and in green for der16. Short deletions are present on both parental chromosomes, 4 nt on chr16 and 5 nt on chr22. On chr16 palindromic sequences are present on each side of the breakpoint. A six nucleotide (nt) insertion is present in the junction of der22 (TTATAC), likely due to template slippage (TSL) copying from the palindrome sequence using a 3 nt (TTA) microhomology. A 2 nt microhomology is present in der16.
(B) Sequence alignment from case 851-06Ö. Derivative chromosome 17 (der17) is shown on top and derivative chromosome X (derX) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr17 and chrX) sequences are labeled in blue for der17 and in green for derX. Short deletions are present on both parental chromosomes, 9 nt on chr17 and 7 nt on chrX. On der17 a 5 nt microhomology is present. A 12 nt insertion is present on der X that may originate from template switching to two different places on chr17 (- strand). In the TS1 a potential CACCT microhomology is present but for TS2 no microhomology could be observed.
(C) Sequence alignment from case 337-01D. Derivative chromosome 3 (der3) is shown on top and derivative chromosome 12 (der12) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr3 and chr12) sequences are labeled in blue for der3 and in green for der12. Deletions are present on both parental chromosomes, 6594 nt on chr3 and 13 nt on chr12. On der3 a 1 nt microhomology is present
A 10 nt insertion is present on der 12 that may have arisen through error prone backward slippage using a 4 nt (TTTT) microhomology.
(D) Sequence alignment from case 29-03E. Derivative chromosome 9 (der9) is shown on top and derivative chromosome 16 (der16) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr9 and chr16) sequences are labeled in blue for der9 and in green for der16. On der 9, 3 nt microhomologies are present on both sides of a 27 nt resection/deletion. On der16 the junction presents with a 222 nt deletion and the 5 bp insertion (TTGGC) originates from inside the deletion.