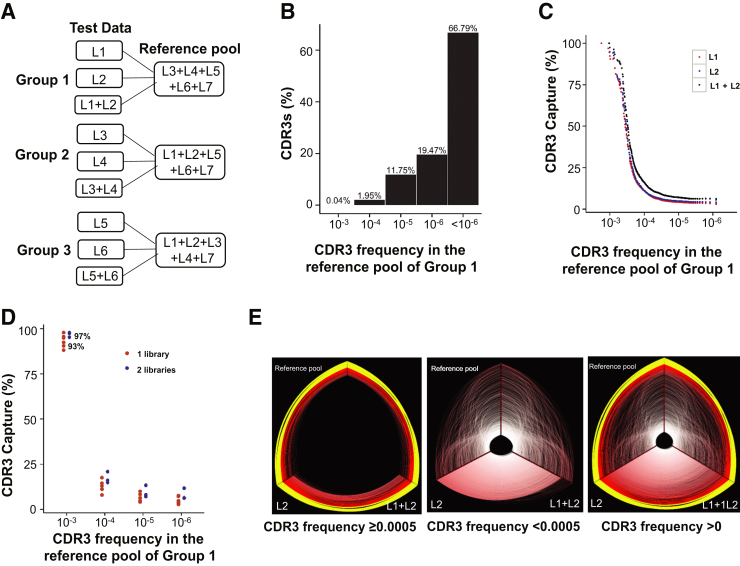
Figure 5.
Capture of CDR3 clones in different biological replicate libraries made from the same genomic DNA stock of a healthy person (HP10). A: Schematic diagram of three analysis groups. Seven different biological replicate libraries were made from seven different PCRs from the same genomic DNA stock and were sequenced separately. Three two-versus-five (two different biological replicate libraries versus the combination of the other five biological replicate libraries) analytical groups were set up from the sequencing data of these seven biological replicate libraries. In each group, the CDR3 clones of two different libraries were individually or jointly compared with those of the other five libraries combined (the reference pool). B: Percentage of CDR3 clones at different ranges of frequency in the reference pool of Group 1. The results for Groups 2 and 3 (data not shown) show a similar pattern. C: Percentage of CDR3s in the reference pool of Group 1 captured by each of two biological replicate libraries (L1 and L2) and the combination of the two biological replicates (L1 + L2). The x axis represents CDR3 frequency in the reference pool. Each dot is the sum of all the CDR3 clones with the same frequency. The results of Groups 2 and 3 (data not shown) display a similar pattern. D: Representation of likelihood of a given library or two replicate libraries containing a clonotype present in the reference pool (the combination of the other five replicate libraries as indicated in A) of a healthy individual, as % capture on the y axis versus clonotype frequency on the x axis. Only higher frequency clones are reliably detected. E: Hive plots of shared CDR3 clonotypes among different biological replicate libraries made from the same DNA stock (Group 1). In each hive plot, the three axes represent the CDR3 clones of L2, the union of L1 and L2, and the reference pool. The yellow arcs indicate the common clonotypes with frequency ≥0.01 on at least one of the two compared axes, and the red arcs are secondary clonotypes with frequency <0.01. Only the top 1000 clones by frequency in the repertoire on each axis are displayed.