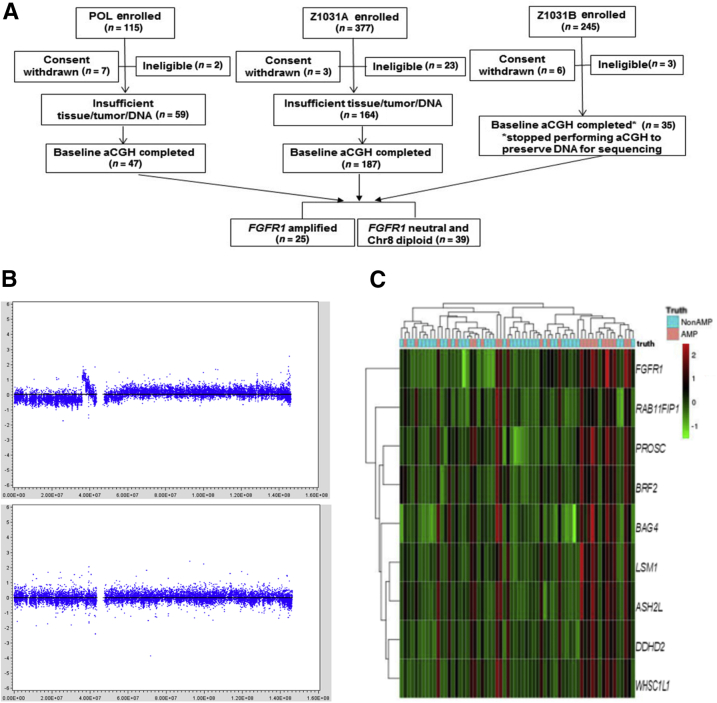
Figure 1.
Identification of genes discriminating FGFR1 amplification from FGFR1 neutral in the discovery cohort [ie, the preoperative letrozole (POL)-Z1031 cohort]. A: The REMARK diagram shows 25 FGFR1-amplified estrogen receptor–positive (ER+) tumors were chosen to be compared against 39 FGFR1 neutral and chromosome 8 diploid ER+ tumors for genes discriminative of FGFR1 amplification. B: Firestorm analysis. The segmented array comparative genomic hybridization (aCGH) copy number signals from the circular binary segmentation algorithm (in log2 ratio, y axis) were plotted against chromosome 8 genome locations (x axis). The top panel shows a FGFR1 firestorm amplified tumor example, and the bottom panel shows a FGFR1 neutral and chromosome 8 diploid tumor example. C: Heat map of the mRNA gene expression levels (in red/green color scheme) of the nine genes (at row). Overexpression of the genes corresponds well to the FGFR1-amplified cases among the 64 samples (at column), as determined by the firestorm analysis (indicated in the top image bar as truth: AMP in pink for amplification and NonAMP in blue for nonamplification).