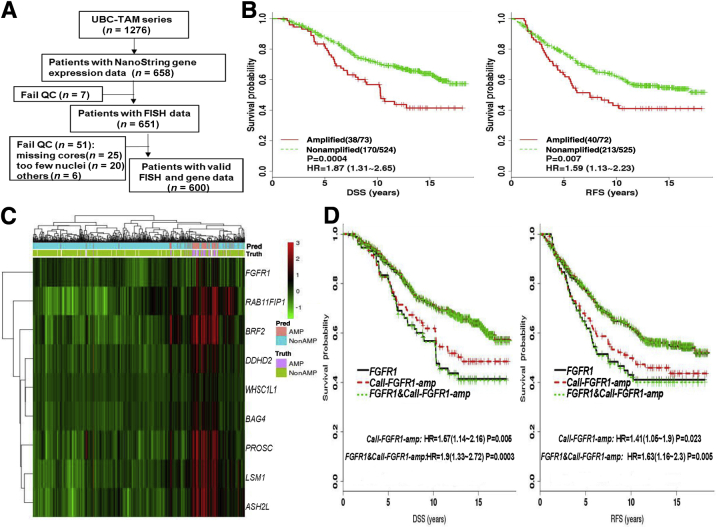
Figure 4.
Validation of FGFR1 amplification calls by Call-FGFR1-amp in the University of British Columbia (UBC)–tamoxifen (TAM) cohort. A: The REMARK diagram; 651 estrogen receptor–positive (ER+) UBC-TAM samples were analyzed for FGFR1 amplification analyses. B: Kaplan-Meier (KM) curves by true FGFR1 amplification status, as determined by fluorescent in situ hybridization (FISH), show prognostic effects for disease-specific survival (DSS) and relapse-free survival (RFS). C: Heat map of the nine selected genes (at row) shows concordance between the genes' overexpression, the amplification statuses called by Call-FGFR1-amp, and the FGFR1 FISH amplification statuses of the 651 ER+ tumors (at column), indicated in the top image bar by pred (for Call-FGFR1-amp) and truth (for FISH), respectively. AMP in pink/purple for amplification and NonAMP in blue for nonamplification. White bars are for cases with nonreadable FISH results. D: Overlaid KM curves exhibit close tracking of Call-FGFR1-amp for FGFR1 FISH, showing similar prognostic effects. KM curves for DSS and RFS by FGFR1 amplification based on Call-FGFR1-amp (Call-FGFR1-amp: red dashed curves, upper/lower red curves corresponding to nonamplified versus amplified patients, as determined by Call-FGFR1-amp, respectively), overlaid with curves by the FISH gold standard (FGFR1: black solid curves, upper/lower curves corresponding to nonamplified versus amplified patients, as determined by FISH, respectively) and FGFR1 status, as determined by both Call-FGFR1-amp and FISH (FGFR1 and Call-FGFR1-amp: green dotted curves, the upper green dotted curve representing patients who were FGFR1 FISH amplified and called so by Call-FGFR1-amp versus the lower green dotted curves representing the remaining patients who were called nonamplified by both or had inconsistent calls between FISH and Call-FGFR1-amp). The hazard ratios (HRs) with 95% CIs and log-rank test P values (P) were indicated in the legend. QC, quality control.