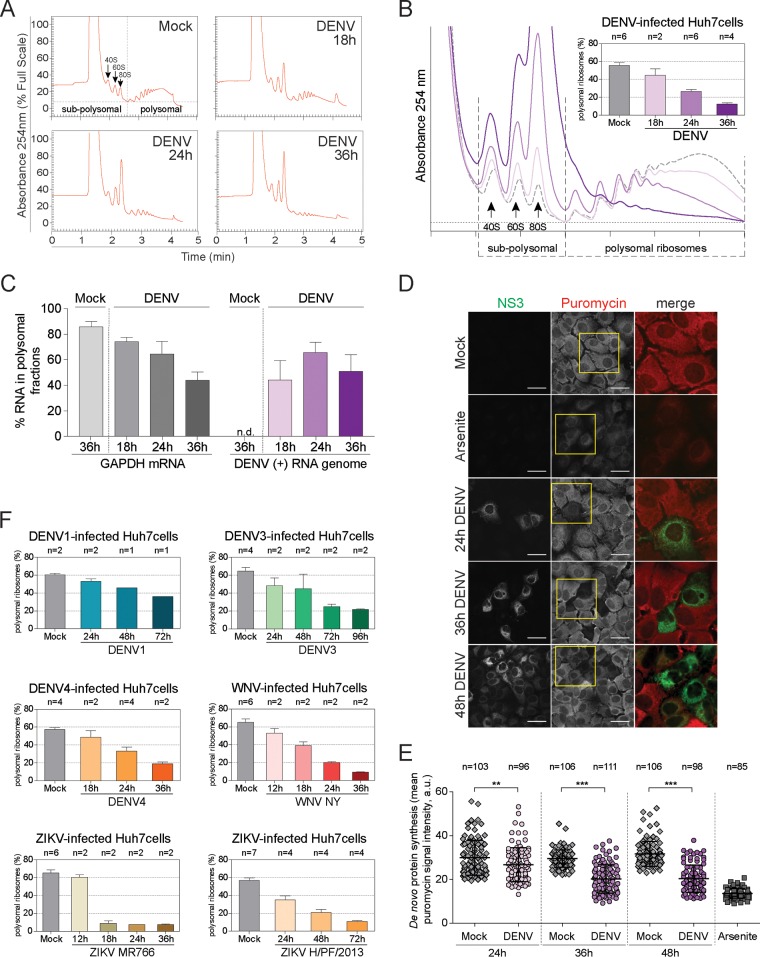
FIG 1.
Induction of host translation repression by flavivirus infection. (A) Representative polysome profiles of naive Huh7 cells (Mock) and DENV-infected cells at 18, 24, and 36 h p.i. Cell extracts of naive Huh7 cells or cells infected with DENV (MOI of 10) for the indicated time periods were loaded on a sucrose gradient and separated by ultracentrifugation. Sucrose gradients were eluted from the top using a fractionator, and absorption at 254 nm was continuously recorded. Shown in the Mock panel is the separation between actively translated, polysomal mRNAs associated with multiple ribosomes and not or poorly translated subpolysomal mRNAs (40S and 60S, single ribosomal subunits; 80S, monosome). DENV-infected cells show an increase of the monosomal 80S peak throughout the infection. (B) Representative polysome profile analysis (lower panel). The percentage of polysomal ribosomes (actively translating mRNAs) is assessed by measuring the area below the polysomal part of the curve and the area of subpolysomal and polysomal parts of the curve. Histogram bars shown in the upper panel represent the mean percentages of polysomal ribosomes ± standard error of the mean (SEM). n, number of profiles analyzed. (C) Abundance of specific mRNAs in gradient fractions. Polysome profiles of naive (Mock) and DENV-infected Huh7 cells were recorded at the indicated time points p.i. Total RNA was extracted from all fractions, and the relative abundance of specific mRNAs in each fraction was quantified by qRT-PCR. Histogram bars represent mean percentages of GAPDH mRNA and DENV positive-strand RNA genome associated with the polysomal fractions ± standard deviation (SD) (n = 3). (D) Reduction of protein synthesis in DENV-infected cells. Naive Huh7 cells (Mock) and cells infected with DENV for the indicated time period were treated with puromycin to induce a premature release of nascent polypeptidic chains. Arsenite-treated Huh7 cells were used as a control. Puromycylated chains are visualized using an antipuromycin antibody (red) and infection by immunostaining of DENV NS3 (green). Representative fields of view are shown. Yellow squares represent the cropped section shown in the merge panel. Scale bars, 50 µm. (E) Scatter plot of de novo protein synthesis measured by fluorescence intensity of the puromycin signal (mean fluorescence intensities ± SD; n = 3) a.u., arbitrary units. Statistical significance and the number of analyzed cells (n) are given at the top. ***, P < 0.001; **, P < 0.01. (F) Host cell translation repression is a general feature of flavivirus infection. Polysomal profiles of Huh7 cells infected with DENV serotype 1, 3, or 4, WNV strain NY, or ZIKV MR766 or H/PF/2013 (MOI of 10) for the indicated time periods were recorded (see Fig. S2 in the supplemental material). Shown are mean percentages of polysomal ribosomes ± SEM. n, number of profiles analyzed.