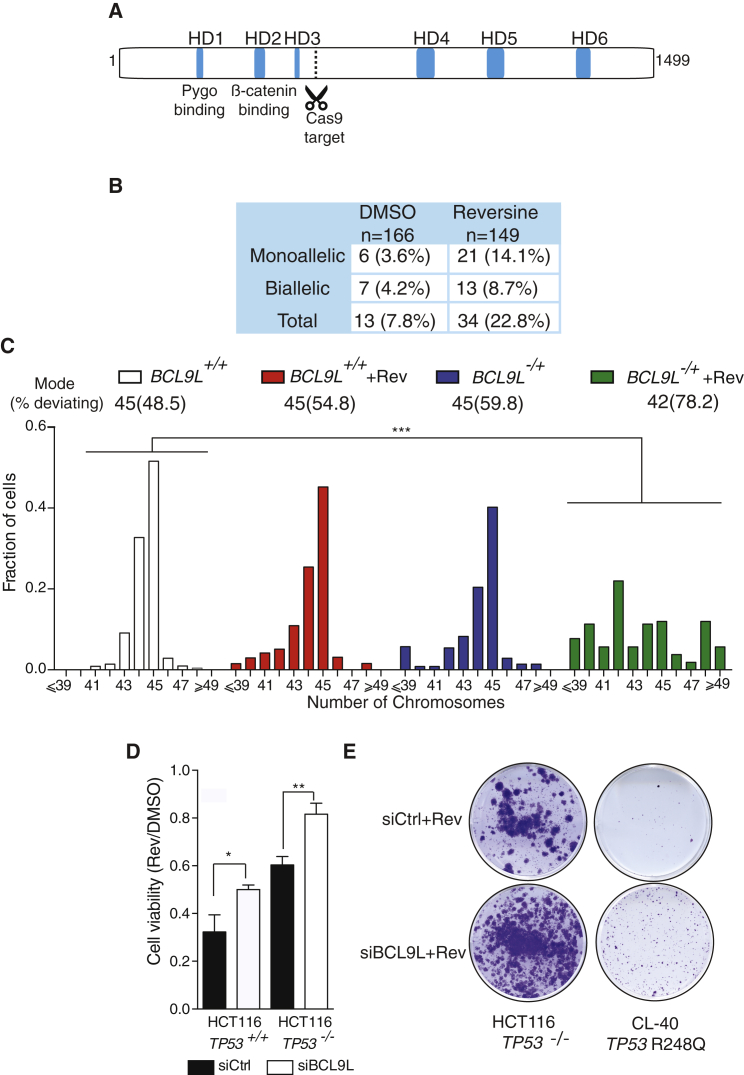
Figure 3.
Heterozygous Truncation of BCL9L Drives Aneuploidy Tolerance in HCT-116 Cells
(A) Mapping of the CRISPR protospacer site on the BCL9L protein. Guide RNA targets nucleotides 2,542–2,561 (BCL9L cDNA sequence GenBank: NM_182557).
(B) Genotyping results of BCL9L CRISPR clones selected in 125 nM reversine for 15 days (percentage of all isolated clones).
(C) Karyotypic analysis of an isolated HCT-116 clone bearing a heterozygous 5-bp deletion C-terminal to the HD3 domain in BCL9L (p.Glu530fs) (70–100 cells analyzed, modal number of chromosomes and percent deviating from the mode shown above the graph; p value calculated by two-sided Wilcoxon rank-sum test).
(D) Effect of BCL9L depletion on cell viability of isogenic TP53-WT and null HCT-116 cells (72 hr, n = 3; p value calculated by paired Student's t test). Cell viability was measured by CellTiter-Blue (Promega).
(E) Colony-formation assay in CRC TP53-mutant cells (TP53-null HCT-116 and CL-40).
Error bars denote SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S5.