Abstract
Background
Geckos of the genus Pachydactylus and their close relatives comprise the most species-rich clade of lizards in sub-Saharan Africa. Many explanations have been offered to explain species richness patterns of clades. In the Pachydactylus group, one possible explanation is a history of diversification via geographic isolation. If geographic isolation has played a key role in facilitating diversification, then we expect species in more species-rich subclades to have smaller ranges than species in less diverse subclades. We also expect traits promoting geographic isolation to be correlated with small geographic ranges. In order to test these expectations, we performed phylogenetic analyses and tested for correlations among body size, habitat choice, range sizes, and diversification rates in the Pachydactylus group.
Results
Both body size and habitat use are inferred to have shifted multiple times across the phylogeny of the Pachydactylus group, with large size and generalist habitat use being ancestral for the group. Geographic range size is correlated with both of these traits. Small-bodied species have more restricted ranges than large-bodied species, and rock-dwelling species have more restricted ranges than either terrestrial or generalist species. Rock-dwelling and small body size are also associated with higher rates of diversification, and subclades retaining ancestral conditions for these traits are less species rich than subclades in which shifts to small body size and rocky habitat use have occurred. The phylogeny also illustrates inadequacies of the current taxonomy of the group.
Conclusions
The results are consistent with a model in which lineages more likely to become geographically isolated diversify to a greater extent, although some patterns also resemble those expected of an adaptive radiation in which ecological divergence acts as a driver of speciation. Therefore, the Pachydactylus group may represent an intermediate between clades in which radiation is adaptive versus those in which it is non-adaptive.
Keywords: Biogeography, Systematics, Timetree, Allopatry, Radiation, Cladogenesis, Ancestral reconstruction, Phylogenetic comparative methods
Background
Discrete geographic regions, both continentally and on islands, often have biotas dominated by a relatively small number of species-rich lineages. The most obvious of these dominant groups are adaptive radiations in which a single ancestral species has given rise to descendants filling numerous niches, examples of which include Galapagos Finches (14 sp., 58% of breeding songbird species in the archipelago), Lake Victoria cichlids (169 sp., 67% of ray-finned fishes in the lake), and West Indies Anolis lizards (168 sp., 38% of lizards native to the islands) [1–3]. However, there are many other less-known examples of regionally prominent radiations. Among lizards, one of the most striking are geckos of the genus Pachydactylus (56 species) and its close relatives Chondrodactylus (6 species), Colopus (2 species), and Elasmodactylus (2 species). By species number, these geckos are the most successful radiation of lizards in southern Africa. Sixty four of 66 species occur in the southern African subcontinent, defined as that part of Africa south of the Zambezi and Kunene rivers, and most are endemic to this region. These species occupy all major habitat types in southern Africa, and many species in the group display morphological novelties such as loss of adhesive toe pads or the evolution of interdigital webbing [4, 5]. Numerous other gecko genera are found in southern Africa, most of which are endemic or reach their peak diversity there, including Afroedura, Afrogecko, Cryptactites, Goggia, Homopholis, Narudasia, Ramigekko, and Rhoptropus, but Pachydactylus group species often dominate the gekkonid fauna, comprising, for example, 13 of 18 species in the Richtersveld of South Africa [6]. Likewise, none of these other genera approach Pachydactylus in its diversity of morphological or ecological variation.
Numerous possible causative factors have been posited or shown to explain the relative success of species-rich organismal groups. In classic adaptive radiations, the rapid evolution of morphological disparity may be the key process in spurring lineage accumulation [7, 8]. In other cases, the evolution of a novel trait may allow organisms possessing that trait to access underutilized resources, with utilization promoting ecological diversification and lineage accumulation. Examples include the evolution of antifreeze proteins in Antarctic icefishes and evolution of the pharyngeal jaw structures in parrotfishes [9, 10]. Sexual selection may also promote lineage accumulation, especially when this selection is for traits that serve as prezygotic isolating mechanisms such as male advertisement calls or color patterns serving as visual mate recognition systems [11, 12]. Finally, in some cases species-rich organismal groups may not actually exhibit a high diversification rate at all, but instead have a longer history of occupancy in a geographic region [13].
In the case of Pachydactylus and its relatives, none of these potential explanations are likely to fully account for the observed species diversity. Pachydactylus is divided into eight well-defined species groups [14, 15], each of which is believed to be monophyletic, and each of which is morphologically conservative. The degree of morphological disparity between these groups is not of a magnitude expected in a classic adaptive radiation [8]. This is not to say that there is no morphological variation within the genus. Some morphological novelties have evolved, including the previously mentioned foot characteristics as well as variation in scalation. However, such morphological novelties do not appear to be strong drivers of speciation in the group. For example, the most species-rich radiation of Pachydactylus geckos that has lost toe pads contains only three species, one of which still has toe pads [4].
Many geckos do have visual display systems or other traits which could theoretically promote diversification via divergent sexual selection. Examples include the semaphore geckos (Pristurus), the dwarf geckos (Sphaerodactylus), and various day geckos (including Cnemaspis, Lygodactylus, and Phelsuma) [16–18]. In these genera, males are boldly patterned and use signaling behaviors to defend territories or attract mates. Pachydactylus and its relatives are strictly nocturnal, however, and typically have a drab pattern. Nor are any other prezygotic isolating mechanisms evident that could plausibly be hypothesized to be under sexual selection. Finally, the relative age of Pachydactylus and its relatives likely does not account for its diversity, either – the next closest relative of the Pachydactylus group is Rhoptropus [5], which also occurs mainly in southern Africa but includes only nine species.
Given that none of these explanations can fully account for the species diversity observed in Pachydactylus and its relatives, we hypothesize that geographic isolation leading to allopatric divergence plays a key role in lineage accumulation in Pachydactylus and its relatives. Populations in allopatry, if isolated for a sufficient period of time, can naturally speciate via genetic drift without requiring significant contributions from natural selection acting on divergent morphological traits or sexual selection promoting differentiation of mating systems among species [19, 20]. If geographic isolation does play a key role in diversification of Pachydactylus and its relatives, then traits promoting the formation of geographic isolation should affect both species’ range sizes and diversification rate. The heritability of range size has been a matter of debate, but increasing numbers of studies demonstrate its heritability [21–25]. In at least some clades, including lizards, this heritability is associated with variable morphological or ecological traits [26, 27]. Likewise, numerous studies have reported trait-associated variation in diversification rates, especially since the development of BiSSE (binary-state speciation and extinction) and related models [28].
For Pachydactylus and its relatives, two variable traits of interest that may promote geographic isolation are body size and habitat preference. There is substantial body size variation in Pachydactylus and related genera, with the largest and smallest species having adult snout–vent lengths of 35 and 113 mm, respectively [29]. In many groups body size has been shown to be positively correlated with range size [30]. Habitat preference within Pachydactylus varies, with species showing preferences ranging from sand dunes to rocky cliffs to houses. In southern Africa, the periodic advance and retreat of Kalahari and Namib sands over geological time is linked to climatic variation [31–33]; this process has likely allowed intermittent connections to form between adjacent rocky habitats, but the prevailing pattern is that terrestrial habitats are relatively continuous while rocky habitats are more discontinuous. As a result, a preference for rocky habitats may be expected to be associated with geographic isolation and smaller range sizes. Such substrate specialization has been suggested to facilitate speciation in the Pachydactylus group [14, 34], but has never been explicitly tested.
We test whether body size or habitat preference is associated with the formation of geographic isolation in the Pachydactylus group in a phylogenetic context. We have generated a comprehensive time-calibrated multi-locus phylogeny of the group, and obtained body size and habitat preference trait data for all ingroup species. Geographic range size estimates are produced for all species, and the association between trait data and range size is quantified. We also estimate patterns of lineage accumulation through time and trait-associated estimates of diversification. Our data show that both body size and habitat preference affect range size, and that variation in these traits is also correlated with variation in diversification rate, suggesting that allopatric divergence following isolation has played a key role in speciation in the Pachydactylus group.
Methods
Phylogeny estimation
Previous studies have confirmed that the Pachydactylus is part of a monophyletic assemblage of morphologically similar geckos, also including genera Chondrodactylus, Colopus, and Elasmodactylus [4, 14, 35]. We sought to estimate a comprehensive phylogeny for this group, and obtained genetic samples from individuals of 55 of 56 Pachydactylus species, 6 of 6 Chondrodactylus, both Colopus species, and both Elasmodactylus species. These genera are part of a larger clade of geckos mainly distributed in Africa and Madagascar, and within this larger grouping they are most closely related to the genus Rhoptropus [5, 36]. As such, we included exemplars of 9 of 9 Rhoptropus species to serve as a near outgroup. An additional 18 gekkotan and 4 non-gekkotan taxa (Anolis, Gallus, Python, Trachylepis) were included as more distant outgroups, with outgroup species choice partially determined based on utility for molecular clock calibration. Nearly all ingroup sequences are associated with vouchered museum specimens. Sequences for four species (Elasmdactylus tuberculosus, Pachydactylus namaquensis, P. tsodiloensis, P. visseri) are exceptions, with sequences derived from genetic material obtained from captive-bred individuals; in these cases the live specimens were viewed by the authors to confirm identification and associated genetic material has been deposited in the Cryogenic Collection at the Museum of Comparative Zoology, Harvard University.
We constructed a sequence data set of nuclear and mitochondrial genes that evolve in a relatively clocklike fashion and have proven useful for determining relationships among species within gekkonid genera [37, 38]. The combined data set is 3443 bp (base pairs), including portions of the nuclear genes RAG1 (recombination activating gene 1; 1053 bp), KIF24 (kinesin family member 24; 592 bp) and PDC (phosducin; 395 bp), along with the complete mitochondrial ND2 gene (NADH dehydrogenase subunit 2; 1041 bp) and several adjacent tRNA genes (transfer RNA; 361 bp) (Table 1). All newly generated sequences were deposited in GenBank (accession numbers KY224166–KY224347).
Table 1.
Specimens and GenBank accession numbers of specimens used in this study
| Species | ID Number | ND2 | Rag1 | PDC | KIF24 |
|---|---|---|---|---|---|
| Anolis carolinensis | n/a | EU747728 | AAWZ 02015549 | AAWZ 02013979 | NW_003338919 |
| Chondrodactylus angulifer | MCZ R-184985 | KY224209 | KY224307 | KY224257 | |
| Chondrodactylus bibronii | CAS 201841 | JN543886 | JN543930 | KY224258 | KY224166 |
| Chondrodactylus fitzsimonsi | MCZ R-185712 | JN393945 | KY224308 | KY224259 | KY224167 |
| Chondrodactylus laevigatus | MCZ R-184819 | KY224211 | KY224310 | KY224260 | KY224168 |
| Chondrodactylus pulitzerae | CAS 193828 | KY224210 | KY224309 | ||
| Chondrodactylus turneri | MCZ R-184410 | KY224249 | KM073525 | KM073612 | KM073800 |
| Coleonyx variegatus | MVZ 161445 | AB114446 | |||
| Coleonyx variegatus | CAS 205334 | EF534777 | EF534817 | ||
| Colopus kochii | CAS 214803 | KY224212 | KY224311 | KY224261 | KY224169 |
| Colopus wahlbergii | NMZB 16974 | JN569158 | JN569191 | JQ945366 | |
| Correlophus ciliatus | AMS R-146595 | JX024438 | EF534778 | EF534818 | KU157544 |
| Elasmodactylus tetensis | PEM R-5540 | KY224213 | KY224312 | KY224262 | KY224170 |
| Elasmodactylus tuberculosus | MCZ:Cryo 3006 | KY224214 | KY224313 | KY224263 | KY224171 |
| Euleptes europaea | no number | JN393941 | EF534806 | EF534848 | KU157420 |
| Gallus gallus | n/a | KT626857 | NM_001031188 | XM_004943303 | NC_006127 |
| Goggia braacki | PEM R-11911 | KM073689 | KM073528 | KM073614 | KM073802 |
| Nephrurus levis | AMS 140561 | GU459544 | GU459746 | KU157421 | |
| Nephrurus levis | SAMA R-19968 | AY369018 | |||
| Oedura marmorata | SAMA R-34209 | AY369015 | |||
| Oedura marmorata | AMS 143861 | EF534779 | EF534819 | KU157428 | |
| Ophidiocephalus taeniatus | SAM R-44653 | AY134601 | HQ426303 | HQ426214 | KU157422 |
| Pachydactylus acuminatus | MCZ R-185739 | KY224215 | KY224314 | KY224264 | KY224172 |
| Pachydactylus affinis | PEM R-17545 | KY224216 | KY224315 | KY224265 | |
| Pachydactylus amoenus | AMB 8670 | JN569163 | |||
| Pachydactylus angolensis | CAS 254887 | KY224217 | KY224316 | ||
| Pachydactylus atorquatus | MCZ R-184811 | KY224218 | KY224317 | KY224266 | |
| Pachydactylus austeni | LSUMZ H1629 | KY224250 | JQ945321 | JQ945389 | KY224173 |
| Pachydactylus barnardi | MCZ R-184749 | KY224219 | KY224318 | KY224267 | |
| Pachydactylus bicolor | NMNW (AMB 7631) | JN543870+ KY224220 | JN543911 | KY224268 | |
| Pachydactylus boehmei | MCZ R-184883 | JN543906 | JN543947 | KY224270 | KY224174 |
| Pachydactylus capensis | MCZ R-184499 | HQ165962 | HQ165992 | HQ165977 | KY224175 |
| Pachydactylus caraculicus | MCZ R-185767 | JN543889 | JN543933 | KY224271 | |
| Pachydactylus carinatus | LSUMZ 57293 | KY224221 | KY224319 | KY224272 | |
| Pachydactylus etultra | MCZ R-184978 | HQ165959 | HQ165989 | HQ165974 | KY224176 |
| Pachydactylus fasciatus | MCZ R-185759 | HQ165949 | HQ165978 | HQ165963 | |
| Pachydactylus formosus | CAS 206715 | KY224222 | KY224320 | KY224273 | |
| Pachydactylus gaiasensis | MCZ R-184169 | JN543891 | KM073533 | KM073615 | KY224177 |
| Pachydactylus geitje | PEM R-11226 | JN543887 | JN543931 | KY224274 | KY224178 |
| Pachydactylus goodi | MCZ R-184783 | KY224223 | KY224321 | KY224275 | KY224179 |
| Pachydactylus griffini | MCZ R-185741 | KY224224 | KY224322 | KY224276 | KY224180 |
| Pachydactylus haackei | CAS 186341 | KY224225 | KY224323 | KY224277 | |
| Pachydactylus kladaroderma | PEM R-1253 | KY224251 | JQ945323 | JQ945391 | |
| Pachydactylus kobosensis | CAS 223904 | KY224226 | KY224324 | KY224278 | KY224181 |
| Pachydactylus labialis | MCZ R-184758 | KY224227 | KY224325 | KY224279 | KY224182 |
| Pachydactylus latirostris | PEM R-16720 | JN569141 | JN569173 | KY224280 | KY224183 |
| Pachydactylus macrolepis | PEM R-17668 | JN569139 | JN569170 | KY224281 | KY224184 |
| Pachydactylus maculatus | CAS 186380 | KY224228 | KY224326 | KY224282 | KY224185 |
| Pachydactylus maraisi | NMNW (JV 1856) | JN543871 | JN543912 | KY224269 | |
| Pachydactylus mariquensis | NMB R10936 | JN569157 | JN569190 | KY224283 | KY224186 |
| Pachydactylus mclachlani | MCZ R-185094 | HQ165950 | HQ165980 | HQ165965 | KY224187 |
| Pachydactylus monicae | CAS 193418 | HQ165952 | HQ165982 | HQ165967 | KY224188 |
| Pachydactylus montanus | MCZ R-184243 | KY224229 | KY224327 | KY224284 | KY224189 |
| Pachydactylus namaquensis | MBUR 01770 | KY224230 | |||
| Pachydactylus namaquensis | MCZ:Cryo 3007 | KY224328 | KY224285 | KY224190 | |
| Pachydactylus oculatus | PEM R-1284 | KY224231 | KY224329 | KY224286 | |
| Pachydactylus oreophilus | MCZ R-185769 | JN543892 | JN543936 | KY224287 | KY224191 |
| Pachydactylus oshaughnessyi | NMZB (DGB 611) | KY224232 | KY224330 | KY224288 | KY224192 |
| Pachydactylus otaviensis | MCZ R-184867 | JN543893 | JN543937 | KY224289 | KY224193 |
| Pachydactylus parascutatus | CAS 214750 | JN543894 | JN543938 | KY224290 | KY224194 |
| Pachydactylus punctatus | PEM R-12461 | KY224233 | KY224331 | KY224291 | |
| Pachydactylus purcelli | PEM R-16895 | HQ165954 | HQ165984 | HQ165969 | |
| Pachydactylus purcelli | MCZ R-184796 | KY224195 | |||
| Pachydactylus rangei | MCZ R-183725 | JN543907 | JN543948 | JQ945392 | |
| Pachydactylus reconditus | MCZ R-184856 | KY224234 | KY224332 | KY224292 | |
| Pachydactylus robertsi | NMNW R6697 | KY224235 | KY224333 | KY224293 | |
| Pachydactylus rugosus | CAS 201905 | KY224252 | JQ945325 | JQ945393 | |
| Pachydactylus sansteynae | CAS 214589 | JN543898 | KY224334 | KY224294 | |
| Pachydactylus scherzi | MCZ R-184938 | KY224236 | KY224335 | KY224295 | |
| Pachydactylus scutatus | MCZ Z37843 | JN543901 | JN543943 | KY224296 | KY224196 |
| Pachydactylus serval | MCZ R-185989 | HQ165956 | HQ165986 | HQ165986 | KY224197 |
| Pachydactylus tigrinus | NMB R10936 | KY224237 | KY224336 | KY224297 | KY224198 |
| Pachydactylus tsodiloensis | MCZ:Cryo 3008 | KY224238 | KY224337 | KY224298 | KY224199 |
| Pachydactylus vansoni | MCZ R-184434 | KY224239 | KY224299 | KY224200 | |
| Pachydactylus vanzyli | NMNW (JV1761) | KY224253 | JQ945326 | JQ945394 | KY224201 |
| Pachydactylus visseri | MCZ:Cryo 3009 | KY224240 | KY224338 | KY224300 | KY224202 |
| Pachydactylus waterbergensis | MCZ R-184751 | KY224241 | KY224339 | KY224301 | |
| Pachydactylus weberi | PEM R-12449 | HQ165960 | HQ165990 | HQ165975 | KY224203 |
| Pachydactylus werneri | MCZ R-184960 | KY224242 | KY224340 | KY224302 | KY224204 |
| Phelsuma inexpectata | JB 56 | JN393939 | JN393983 | JN394016 | |
| Phelsuma rosagularis | n/a | EU423292 | |||
| Phelsuma rosagularis | JB 109 | HQ426306 | HQ426217 | ||
| Phyllopezus pollicaris | MZUSP 92491 | JX041417 | EU293635 | ||
| Phyllopezus pollicaris | CENPAT12084 | JQ827509 | |||
| Phyllopezus pollicaris | JFBM 15822 | HQ426225 | |||
| Pygopus nigriceps | ERP R29509 | AY134604 | |||
| Pygopus nigriceps | SAMA R-23908 | FJ571628 | |||
| Pygopus nigriceps | MVZ 197233 | EF534823 | |||
| Python bivittatus | n/a | AEQU 010344888 | AEQU 01027927 | NW_006537073 | |
| Python regius | n/a | AB177878 | |||
| Rhoptropus afer | MCZ R-183711 | KY224254 | KM073535 | KM073616 | KM073806 |
| Rhoptropus barnardi | CAS 214658 | KY224243 | KY224341 | KY224303 | KY224205 |
| Rhoptropus benguelensis | ANG_WC1834 | KY224246 | KY224346 | ||
| Rhoptropus biporosus | CAS 224030 | KY224244 | KY224342 | KY224304 | KY224206 |
| Rhoptropus boultoni | CAS 214713 | KY224256 | EF534810 | EF534852 | KY224207 |
| Rhoptropus bradfieldi | NMNW (to be accessioned) | KY224245 | KY224343 | KY224305 | |
| Rhoptropus diporus | MCZ R-183737 | KY224255 | KY224344 | KY224306 | KY224208 |
| Rhoptropus montanus | CAS 254867 | KY224247 | KY224345 | ||
| Rhoptropus taeniostictus | CAS 254908 | KY224248 | KY224347 | ||
| Sphaerodactylus nicholsi | CAS 198444 | KU158020 | EF534786 | EF534826 | KU157415 |
| Sphaerodactylus roosevelti | CAS 198428 | JN393943 | EF534785 | EF534825 | |
| Sphaerodactylus torrei | JB 34 | JX440519 | EF534788 | EF534829 | KU157416 |
| Teratoscincus microlepis | JFBM 15 | KU157417 | |||
| Teratoscincus microlepis | TG 00074 | JX041451 | EF534800 | EF534842 | |
| Teratoscincus roborowskii | CAS 171203 | AF114252 | |||
| Teratoscincus roborowskii | TG 00070 | EF534799 | EF534841 | ||
| Teratoscincus roborowskii | JFBM 14 | KU157418 | |||
| Teratoscincus scincus | CAS 228808 | KU157419 | |||
| Teratoscincus scincus | JFBM 14252 | JX041454 | EF534801 | EF534843 | |
| Trachylepis varia | MCZ-R 184873 | GU931671 | |||
| Trachylepis varia | TNHC 68769 | GU931603 | GU931534 | ||
| Trachylepis varia | ZFMK 68413 | KC345241 | |||
| Woodworthia maculata | RAH 292 | GU459852 | GU459449 | GU459651 | |
| Woodworthia maculata | RAH 92 | KU157432 |
Specimen ID codes are as follows: AMB (Aaron M. Bauer field collection); AMS (Australian Museum, Sydney); ANG_WC (Werner Conradie field collection); CAS (California Acedemy of Sciences); CENPAT (Centro Nacional Patagónico, Puerto Madryn); DGB (Donald G. Broadley field collection); ERP (Eric R. Pianka field collection); JB (Jon Boone tissue collection); JFBM (James Ford Bell Museum of Natural History, University of Minnesota); JV (Jens Vindum field collection); LSUMZ (Louisiana State University Museum of Zoology); MBUR (Marius Burger field collection); MCZ (Museum of Comparative Zoology, Harvard University); MVZ (Museum of Vertebrate Zoology, University of California); MZUSP (Museum of Zoology, University of Sao Paulo); NMB (National Museum, Bloemfontein); NMNW (National Museum of Namibia, Windhoek); NMZB (National Museum of Zimbabwe, Bulawayo); PEM (Port Elizabeth Museum); RAH (Rod A. Hitchmough tissue collection); SAM/SAMA (South Australian Museum); TG (Tony Gamble tissue collection); TNHC (Texas Natural History Collection); ZFMK (Zoologisches Forschungsmuseum Alexander Koenig)
For new sequences generated in this study, DNA was obtained from frozen or ethanol-preserved tissue samples using Qiagen DNeasy tissue kits under the manufacturer’s protocol. PCR (polymerase chain reaction) amplification of fragments was performed in 25 μL reactions, under standard reaction conditions [39]. ND2, tRNA, RAG1, and PDC primers used in PCR and sequencing were the same as those used in [37]; KIF24 primers were derived from [40]. PCR purification was performed using AMPure magnetic beads, followed by cycle sequencing and purification using CleanSeq magnetic beads. Capillary electrophoresis was performed on an Applied Biosystems 3730xl sequencer. Sequence assembly was performed using BioEdit [41] or Geneious 5.1 [42], with alignment using Clustal [43]. Alignments of the protein-coding genes were edited manually to preserve reading frame and checked to ensure absence of premature stop codons, while those of the tRNAs were edited manually to preserve secondary structural features estimated in ARWEN [44].
Phylogenetic analyses were performed using maximum likelihood (ML) and Bayesian (BI) optimality criteria. For each analysis, model and partition choices were separately identified under the Bayesian Information Criterion using PartitionFinder [45]. In each case considered models of evolution were limited to those models that can be implemented by the programs used for phylogeny estimation. Greedy search schemes were employed and thirteen potential data blocks were considered: twelve data blocks corresponding to the three codon positions for each of the four protein-coding genes and the tRNA data comprising the thirteenth data block.
The ML analysis was performed using RAxML 8.2.4 [46]. One hundred independent searches were implemented on the original data set to identify the best tree, followed by 1,000 non-parametric bootstrap replicates to assess branch support. Based on the PartitionFinder results, the data were divided into eight partitions, each using one of two models: ND2 codon position 1, ND2 codon position 2, tRNAs, and (PDC position 1 + 2 + RAG1 position 1 + 2) used the GTR (general time reversible) + I + Γ model, while ND2 position 3, (PDC position 3 + RAG1 position 3), (KIF24 position 1 + 2) and KIF24 position 3 used the GTR + Γ model.
The BI analysis was implemented in BEAST 1.8.2 [47], using a Yule tree prior and uncorrelated lognormal relaxed clock. Based on the PartitionFinder results, the data were divided into ten partitions employing six distinct models: ND2 position 1, ND2 position 2, and tRNAs used the GTR + I + Γ model. ND2 position 3 used the GTR + Γ model. RAG1 position 1 + 2 used the TrN (Tamura-Nei) + I + Γ model. RAG1 position 3 and KIF24 position 1 + 2 used the HKY (Hasegawa-Kishino-Yano) + Γ model. PDC position 3 and KIF24 position 3 used the K80 (Kishino 1980) + Γ model. PDC position 1 + 2 used the TrNef + I + Γ model. Four replicate analyses were run for 50 million generations, sampled every 1000 generations. The first 5 million generations were discarded as burn-in. Effective sample sizes were estimated in Tracer 1.5 (>300 for all parameters in each run) to confirm the chain length was adequate.
BEAST 1.8.2 was also used to estimate divergence times simultaneously with phylogenetic relationships. The root prior (Lepidosauria-Archosauria divergence) was given a normal distribution (mean = 275 Ma [million years ago], SD = 15) encompassing the range of estimates for this divergence [48, 49]. Five constraints were also applied to internal nodes: most recent common ancestor (MRCA) of Phelsuma rosagularis and P. inexpectata (uniform prior; 0–8 Ma; [37]). MRCA of sampled Sphaerodactylus – S. ocoae, S. roosevelti, and S. torrei (exponential prior; mean = 3, offset = 15 Ma; [50, 51]). MRCA of Woodworthia maculata and Oedura marmorata (exponential prior; mean = 17, offset = 16; [52]). MRCA of Ophidocephalus taeniatus and Pygopus nigriceps (exponential prior; mean = 10, offset = 20; [53, 54]). MRCA of Teratoscincus roborowskii and T. scincus (exponential prior; mean = 3, offset = 10; [55]).
Trait data
Body size and habitat preference data were assigned to each species based on the authors’ observations of specimens in the wild (62 of 66 ingroup species have been observed in-situ by the authors), supplemented by examination of vouchered museum specimens and information obtained from the literature [29, 56–58]. Maximum body size was treated in two ways depending on analysis. When possible, SVL (snout-vent length) was treated as a continuous character and the log-transformed maximum SVL was used. However, when treatment of size as a continuous character was not computationally feasible we instead treated size as a binary character. In Pachydactylus and related genera SVL is bimodal (Fig. 1). Those species with a maximum snout–vent length (SVL) < 70 mm comprised the “small” category, and those with a maximum SVL >75 mm comprised the “large” category. Habitat preference was divided into three categories. Those species that primarily shelter in burrows or under surface debris (logs, loose stones, aloe leaves, etc.), and forage actively on the ground, were classified as “terrestrial.” Species that primarily shelter in rock cracks and forage on cliff faces or boulders were classified as “rupicolous.” Finally, unspecialized species that both shelter and forage on a variety of surfaces (rock faces, tree trunks, buildings, etc.) were classified as “generalist climbers.”
Fig. 1.
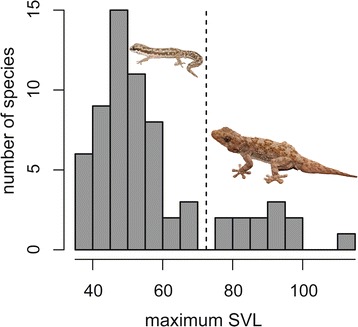
Maximum snout–vent length for species in the genera Pachydactylus, Chondrodactylus, Colopus, and Elasmodactylus. Values are based on literature sources along with observations of field-collected and museum-preserved specimens. The small-bodied species P. geitje and large-bodied P. namaquensis are illustrated
Range size estimates
Extent of occurrence (EOO) and area of occupancy (AOO) were defined as per the current International Union for the Conservation of Nature (IUCN) standards [1]. EOO was calculated as the area of the minimum convex polygon enclosing distribution records for each respective taxon. AOO was initially calculated as the sum of the total area of the quarter degree grid squares within which at least one record occurs. The final AOO was adjusted to an estimate of the actual suitable habitat within the occupied quarter degree squares based on the literature and the authors’ field knowledge of each species. For all endemic South African species and for most species with a portion of their distribution occurring in South Africa, EOO and AOO values were previously estimated as part of the red list evaluation carried out in association with the Atlas and Red List of the Reptiles of South Africa, Lesotho and Swaziland [58]. Calculated EOO usually provides the broadest possible interpretation of the space used by a species, whereas the AOO represents a quite conservative estimate. However, for taxa known from single localities or several localities that are very close to one another, AOO as calculated above may yield a greater area than EOO. We used EOO or AOO, which ever was the greater, as our estimates of species’ ranges. These values were log transformed when used in analyses.
Phylogenetic comparative methods
We performed a variety of comparative analyses to investigate the relationship among phylogeny, divergence times, trait data, and range sizes. All comparative analyses were completed in replicate on both the BEAST maximum clade-credibility tree and on 1000 post-burnin trees randomly sampled from the BEAST posterior distribution. These trees were pruned to remove outgroups (for which we have incomplete taxon sampling and no trait data). The package Phytools [59], implemented in R 3.2.2 [60] was used compute phylogenetic signal of range size using both the K and λ statistics [61, 62]. We also used phytools to plot lineages through time and test for constancy of lineage accumulation through time using the γ statistic of Pybus and Harvey [63].
State dependent diversification of range size on trait data (SVL or habitat) were tested using OUwie [64], which allows for tests of correlation between a multistate vs. continuous trait in a phylogenetic context. Because body size evolution and habitat choice may be coupled [65], we also tested for auto-correlation between these two traits, for a total of three analyses: (1) SVL vs. range size, (2) habitat vs. range size, and (3) SVL vs. habitat. SVL was treated as a multistate (binary) trait in test (1), but was treated as a continuous trait in test (3) to facilitate analysis. In all three cases, we tested the hypothesis that the optimum continuous trait value, θ, differed depending on the identity of the multistate trait value, i.e. whether large- and small-bodied species differ in range size (test 1), whether species differing in habitat use differ in range size (test 2), or whether species differing in habitat use differ in body size (test 3). We performed these tests by estimating ancestral states of the multistate character on the phylogeny, and then fitting values of θ (trait optimum), α (pull toward optimum), and σ (rate of change of trait) for the continuous trait under two model regimes. The null Brownian Motion (BM) model regime estimated single values of θ, α, and σ that did not depend on the state of the multistate character. This was tested against a more complex Ornstein-Uhlenbeck (OU) model in which there were multiple θ parameters, one per multistate character state. We used ΔAICc (corrected Akaike Information Criterion) values to identify which model provided a better fit for the data. All three tests were performed on 1000 trees randomly sampled from the post-burnin BEAST posterior distribution, and on each of these 1000 trees we performed 100 ancestral reconstructions of the multistate trait using stochastic character mapping [66] implemented in phytools, resulting in a total of 100,000 model fits per test, each with a unique combination of phylogeny and ancestral state estimate.
We also estimated trait-associated rates of speciation (λ), extinction (μ), and transition rate (q) under a BiSSE [28] model for SVL data or a multiple-state speciation and extinction (MuSSE) [67] model for habitat data implemented in Diversitree. Because hypothesis testing in a BiSSE or MuSSE framework can have a high Type I error rate [68–70] and low statistical power when data sets contain fewer than several hundred terminal taxa [71], we refrain from explicitly testing the statistical significance of character-associated variation in model parameter estimates. Instead, we fit models in which each trait was given individual λ, μ, and q parameters strictly to determine estimates of these model parameters. Model fitting was performed in an Markov chain Monte Carlo (MCMC) framework with runs lasting 1100 generations and the first 100 discarded as burn-in. These estimates were obtained for each of 1000 trees randomly sampled from the BEAST posterior distribution, resulting in each parameter estimate being obtained from 1,000,000 observations.
Results
Phylogeny and divergence times
The phylogenies estimated in both the ML and BI analyses are very similar (Fig. 2), and most branches receive strong support. As expected, the grouping of Pachydactylus, Chondrodactylus, Colopus, and Elasmodactylus is monophyletic (BI/ML support values 1.0/97) and these are in turn most closely related to Rhoptropus (support values 100/1.0). Within the ingroup, the topology resembles that estimated by Bauer and Lamb [14], which included 26 fewer ingroup taxa and was estimated from ~1,600 fewer nucleotide sites, but there are some notable differences. Most notably, both the genera Colopus and Elasmodactylus are recovered as non-monophyletic. One species of Colopus, C. kochi, is embedded in Pachydactylus and is most closely related to the Pachydactylus mariquensis group (support values 1.0/93), a set of four species represented by a single taxon in [14]. The other Colopus species, C. wahlbergii, is also embedded in Pachydactylus, but there is not strong support for any set of Pachydactylus species being its closest relatives (support values 0.76/54), although there is strong support for its association with Pachydactylus to the exclusion of Chondrodactylus and Elasmodactylus (100/1.0). The two Elasmodactylus species are outside a group containing all Pachydactylus, Colopus, and Chondrodactylus species, with E. tuberculosus being more closely related, but with poor support (0.37/46). Within Pachydactylus, recognized species groups [14, 15, 35, 72–74] are recovered as monophyletic with strong support as are many of the species-level relationships within these groups. However, within the speciose serval/weberi and northwestern groups, in which many new taxa have been added, species relationships are more highly modified. In the first of these, the basal division into reciprocally monophyletic serval and weberi groups is not supported, and the former makes the latter paraphyletic. Relationships among species groups in Pachydactylus remain unresolved, with most groups connected by exceptionally short internodes. There are two exceptions. The serval/weberi group and capensis group are closest relatives, as are the geitje and rugosus groups.
Fig. 2.
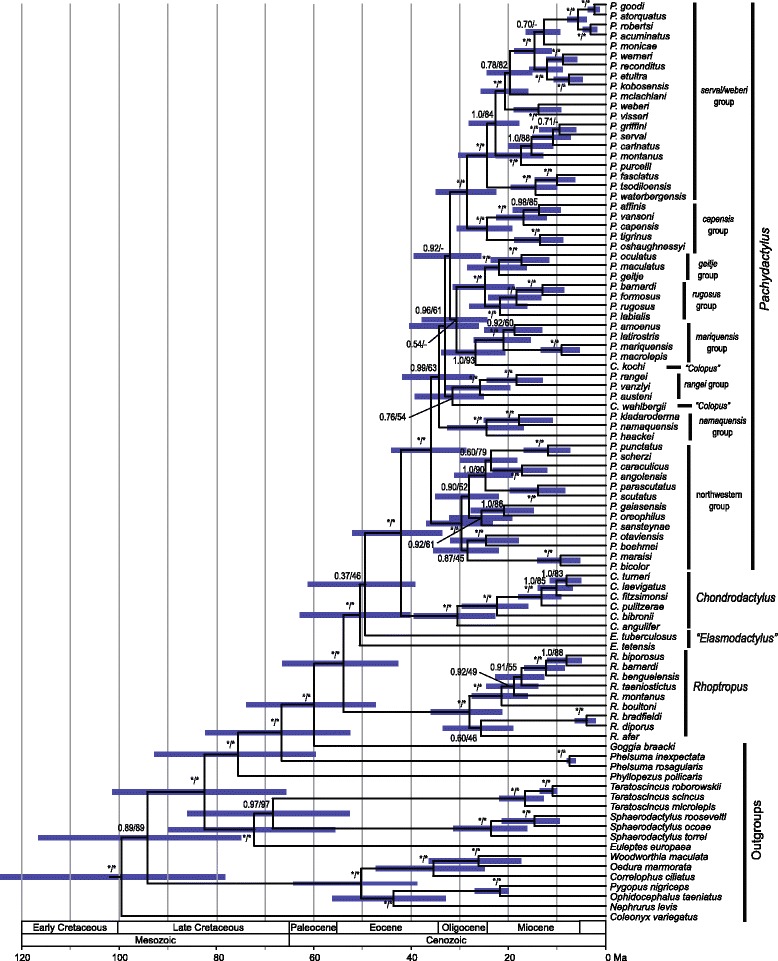
Time-calibrated phylogeny of Pachydactylus and related genera. The topology is the maximum clade credibility tree estimated in BEAST with non-gekkotan outgroups cropped for clarity. Support values (Bayesian posterior probabilities/ML bootstrap) are given at nodes; asterisks indicate nodes with Bayesian support values = 1.0 and ML bootstrap values > 95. Named species groups and genera are given to the right. Geologic epochs and eras are indicated on the timescale; post-Miocene epochs (Pliocene, Pleistocene, Holocene) are not labeled
The divergences between Rhoptropus and Pachydactylus + Chondrodactylus + Colopus + Elasmodactylus occurred in the early Cenozoic (66–43 Ma). This is a similar pattern as observed in other gekkonids, in which relatively species-rich regional radiations undergo initial diversification in the early Cenozoic (e.g. [5, 37, 75]). The short internodes connecting Pachydactylus species groups are indicative of a relatively high diversification rate in the mid-Cenozoic ~30–35 Ma. The lineage through time (LTT) plot shows that the rate of lineage accumulation remains steady or slowly increases to this point after which there is a noticeable decline (Fig. 3). The overall trend is of significantly decreasing lineage accumulation through time (mean γ value = −5.8, p < 1 x 10−5 for all 1000 sampled trees).
Fig. 3.
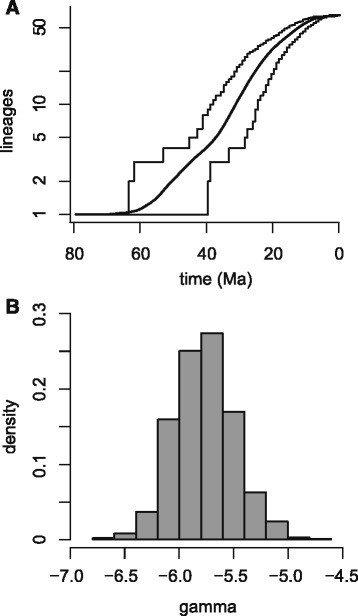
a Lineage accumulation in the Pachydactylus group. The plot depicts LTT curves for 1000 trees randomly sampled from the BEAST posterior distribution. b Histogram of γ statistic estimates for the 1000 LTT curves depicted in (a)
Comparative analyses
Ancestral reconstruction of body size in the Pachydactylus group suggests that being large-bodied is ancestral for the group (Fig. 4). A shift to small body size occurred once early in the evolutionary history of the group, and there have been only two reversals. Reconstruction of habitat preference is more equivocal, but the common ancestor of the group is most commonly reconstructed as a generalized climber (in 80% of reconstructions). What is clear is that more shifts in habitat preference have occurred than shifts in body size, with approximately 26 transitions indicated in total, most commonly between rock-dwelling and terrestrial habitat preferences. Although both habitat and body size are estimated to have shifted multiple times, including reversals, correlation between the two traits is not particularly strong based on fits of BM and OU models — out of 100,000 model fits, the BM model incorporating only a single global SVL optimum was favored according to the AIC 31% of the time (Fig. 5a).
Fig. 4.
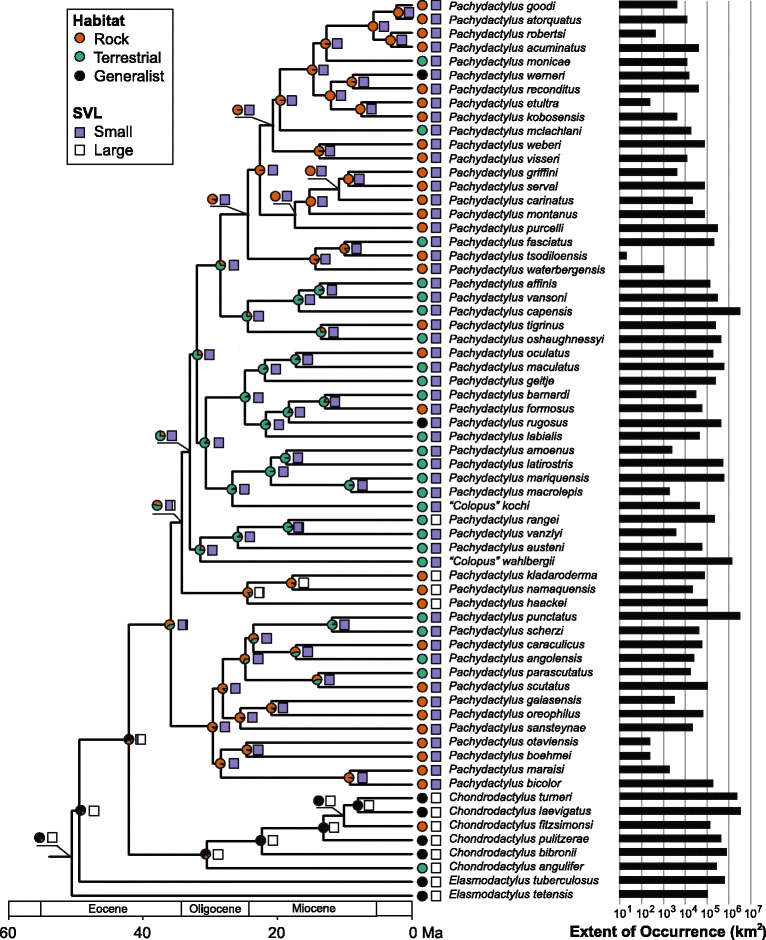
Ancestral states for body size and habitat preference, based on 100 stochastic character maps for each trait on the maximum clade credibility tree. Range size values for each species are given to the right of each terminal branch
Fig. 5.
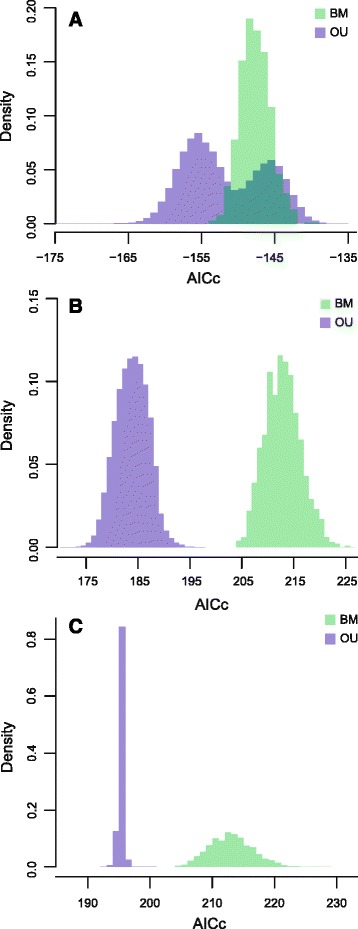
Histogram of AICc values for model fits of Brownian motion (BM) and Ornstein-Uhlenbeck (OU) models of trait diversification estimated in OUwie. a habitat preference vs. SVL. b habitat preference vs. range size. c SVL vs. range size
In contrast, both body size and habitat preference are strongly correlated with range size (Fig. 5b, c). Range size displays significant phylogenetic signal based on Pagel’s λ (λ = 0.46, p = 0.13), but the estimate of K is slightly non-significant (K = 0.57, p = 0.077). The estimated global optimum range size (θ) for small-bodied species is 104.5 km2, approximately one order of magnitude smaller than large-bodied species (θ = 105.4 km2). When comparing habitat preference, rock-dwelling species have the smallest estimated global optimum extent of occurrence (θ = 104.1 km2), followed by terrestrial species (θ = 105.1 km2) with generalized climbers having the largest geographic ranges (θ = 105.8 km2). Trait-associated estimates of speciation and extinction rates are less variable (Fig. 6). Small-bodied species are estimated to have slightly higher speciation (mean λ[small-bodied] = 0.055; mean λ[large-bodied] = 0.040) and lower extinction rates, but there is extensive overlap. Habitat-associated estimates of diversification rate also overlap, especially between terrestrial species and generalized climbers, although rock-dwelling species are estimated to have speciated at somewhat higher rates (mean λ[generalized climber] = 0.032; mean λ[terrestrial] = 0.012; mean λ[rock-dwelling] = 0.065).
Fig. 6.
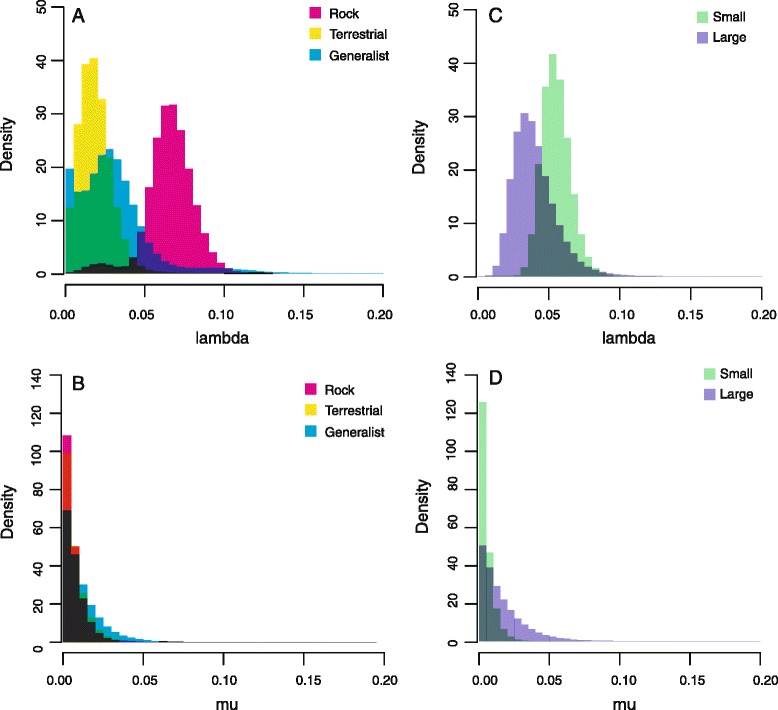
Trait-associated estimates of speciation and extinction generated using Diversitree. Intermediate colors indicate overlap. a habitat-associated speciation rate. b habitat-associated extinction rate. c body size-associated speciation rate. d body-size associated extinction rate
Discussion
While the heritability of range size has been demonstrated for many lineages, possible mechanistic explanations have varied, and include niche breadth [27], dispersal ability [76, 77], and morphological characteristics [26, 78] of lineages, as well as the geographic limits of biomes, landmasses, or hydrological basins [79]. In many cases, these factors may be interlinked. In this study, we focus on two traits, body size and habitat requirements, that were expected to affect dispersal ability either directly because smaller organisms, including some lizards, may disperse shorter distances [80], or indirectly, because habitat patchiness can restrict dispersal if appropriate dispersal corridors are not available [81]. As expected, within the Pachydactylus group the smaller-bodied species occupying more patchily distributed habitats are the species with the smallest geographic ranges. Other studies that have measured dispersal ability directly have shown that reduced dispersal ability does not always lead to reduced range size [77], but in Pachydactylus and its relatives our data suggest that dispersal ability and range size are correlated. Traits affecting dispersal ability are likely not the only factors affecting range size, however. Minimally, it is likely that geographic barriers, including major river systems and mountain ranges, also play a significant role in restricting the ranges occupied by individual species. For example, the species P. austeni and P. goodi are known only from south of the Orange River even though suitable habitats for each of these species also exist to the north [58].
Taken as a whole, the observed patterns of trait evolution, range size, and diversification are consistent with an evolutionary scenario in which diversification has been dominated by geographic isolation followed by allopatric speciation. Based on our analyses, we suggest that geographic isolation has developed more easily in Pachydactylus + Colopus than it has in Elasmodactylus or Chondrodactylus, at least partly as a result of Pachydactylus + Colopus species being more likely to have traits promoting this isolation. Ancestral species in the Pachydactylus group as a whole were most likely large-bodied habitat generalists, and most Chondrodactylus and Elasmodactylus species have retained these traits to the present. We infer small body size and habitat specialization (for either terrestrial or rock-dwelling lifestyles) to appear in the common ancestor of Pachydactylus + Colopus, coincident with a brief observed increase in the rate of lineage accumulation in the Pachydactylus group, followed by a general decline in diversification rate measured across the Pachydactylus group as a whole. Rock-dwelling species especially differ strikingly in range size and diversification rate, having extents of occurrence two orders of magnitude smaller than habitat generalists and estimated rates of diversification 2–4X higher than other species. Allopatric speciation of isolated small-bodied, rock-dwelling lineages therefore can account for much of the observed taxonomic diversity in the Pachydactylus group. Not surprisingly, the subclades that have retained ancestral traits (Chondrodactylus and Elasmodactylus) are much less species-rich than those that have not.
The overall decline in diversification rate through time that we observe in the Pachydactylus group is similar to patterns documented in many lineages that are often attributed to reduced ecological opportunity through time as niches are filled (e.g. [82–85]). In the case of the Pachydactylus group, a general pattern of morphological conservatism within species groups, exemplified by the small number of shifts in body size (Fig. 2) and digital morphology [4, 5] through time is in line with expectations if ecological opportunity has decreased through time. However, shifts in habitat use are more frequent, and the number of co-occurring Pachydactylus group species varies from 1 to 13, suggesting that ecological niche space has not been exhausted. An alternative explanation that may also partly explain the observed rate slowdown is a geographic model as described above. In clades dominated by allopatric speciation, diversification rates may decline as vicariance events affect fewer species as species’ geographic ranges decline through time [86, 87]. The relatively low species diversity of Chondrodactylus, which includes mostly large-bodied habitat generalists (i.e., species with large geographic ranges), compared to Pachydactylus, which includes mostly small-bodied habitat specialists, supports this model.
As indicated above, a jump in lineage accumulation coincident with the appearance of habitat-specialist clades in the mid-Cenozoic ~30–35 Ma is contrary to the general pattern of declining diversification rate through time. It is possible that climatic or geomorphic processes active at this time were especially favorable for isolating lineages, resulting in increased speciation. Major periods of tectonic uplift in eastern and southern Africa did not commence until approximately the Oligocene-Miocene boundary (23 Ma) [88–90], making large-scale geomorphological change incompatible with the observed rate increase. However, a major climatic regime shift did occur at the Eocene-Oligocene boundary: a global cooling associated with Antarctic glaciation [91, 92]. In Africa, this shift resulted in aridification and greater environmental heterogeneity, including reduction in forest cover [93, 94], and would have greatly increased the available habitat for arid-adapted Pachydactylus group geckos, potentially facilitating rapid radiation. A similar pattern occurs in forest-adapted chameleons, where rapid radiation is coincident with wide availability of suitable habitat, in the case of chameleons during the Eocene [95].
In performing this study, we have attempted to minimize confounding factors and metholological biases. For example, we collected data for nearly all target taxa and integrated all comparative analyses across a sample of 1000 credible trees to avoid sampling or phylogenetic biases. Even so, our interpretations should be treated cautiously. The observed relationships between trait data and geographic range extent are based on correlation. While we chose to focus on body size and habitat use specifically because we expected them to affect geographic range, it is possible that one or more other factors co-varying with body size and habitat use are the actual drivers of range size variation among species. Trait-associated measurements of diversification rate utilized the BiSSE model. Even though our ingroup phylogeny was comprehensive, the number of taxa in our data set may not have been large enough to avoid inadequacies of the model [69, 71], which is why we refrain from ascribing statistical significance to these results. Alternate methods of trait-dependant diversification (e.g. [70]) likewise are best suited to larger data sets. One possible way to increase data set size is to incorporate taxa from across the Afro-Malagasy clade of geckos, but the interrelationships of genera within this large radiation are still poorly resolved and relevant trait data are missing for many species. Finally, range size estimates are based on known collection localities and a correct interpretation of species-level taxonomy in the group. Collecting effort varies greatly by country, with, for example, less than 10,000 amphibian and reptile collection records in Angola, 38,000 in Namibia, and >100,000 in South Africa [58, 96]. Some species also vary phenotypically and have named subspecies that further study may reveal to warrant specific status (e.g., Pachydactylus punctatus; [97]). However, given that trait-associations with range size varied by orders of magnitude, we do not expect refinement of species’ range limits or taxonomy to strongly influence our results.
Beyond interpretation of evolutionary patterns, the results of this study also have significant implications for taxonomy and conservation. The phylogenetic results indicate that Elasmodactylus and Colopus are not monophyletic, and both species of Colopus are nested in Pachydactylus. Although we recover Elasmodactylus as non-monophyletic, its monophyly cannot be wholly discounted given the poor support for the node joining E. tuberculosus with Chondrodactylus + Colopus + Pachydactylus. Performing a Shimodaira-Hasegawa (SH) test also shows that the likelihood of our best-scoring tree (lnL −97035.168404) is not significantly higher than the likelihood of a tree in which Elasmodactylus is constrained to be monophyletic (lnL −97036.513963; p > 0.05). These species also share morphological traits rare or absent in other Pachydactylus group species, including preanal pores and easily broken skin [14]. Thus, we suggest that taxonomic decisions regarding these species be delayed until each species’ phylogenetic position is better established. We refer both Colopus species to Pachydactylus. Colopus wahlbergii is morphologically divergent and in our analyses its position in Pachydactylus is equivocal, with moderate support for an association with the rangei group. Thus, we refer it to no species group within Pachydactylus. In contrast, Colophs kochi is deeply nested in Pachydactylus, and there is strong support for its placement as closely related to the P. mariquensis group. This species was also included in Pachydactylus until recently [14]. We therefore advise that C. kochi be re-assigned to the mariquensis group within Pachydactylus.
One important determinant of rarity is range size, and small range size is a key predictor of extinction risk [98, 99]. Thus, species in the Pachydactylus group inheriting traits promoting smaller ranges also inherit traits promoting greater rarity. However, our analyses show these same traits to be associated with higher rates of diversification. Given the difficulty in estimating extinction rates from phylogenies [100], it is unclear if this higher diversification rate is observed despite a higher extinction rate, or if extinction rates do not depend on the measured traits in the Pachydactylus group. Notwithstanding this difficulty, these results stress the importance of defining a frame of reference when measuring evolutionary “success.” In the case of the Pachydactylus group, more widespread, common species tend to belong to relatively species-poor subclades.
Conclusions
The relationships among morphological and ecological traits, range size, and diversification that we observe in the Pachydactylus group points to a history of geographic isolation contributing significantly to the group’s species richness compared to other African geckos. Even so, some aspects of diversification in the Pachydactylus group, including early evolution of divergent traits within the group, are consistent with patterns observed in classic adaptive radiations. In this sense, the process of diversification of Pachydactylus group geckos may be considered intermediate between a true adaptive radiation on one hand and a non-adaptive radiation (as observed in plethodontid salamanders; [101, 102]) on the other. It is likely that many other species-rich groups share this same intermediate pattern.
Acknowledgements
We thank the late Donald G. Broadley, Mirko Barts, Jon Boone, William R. Branch, Marius Burger, and Krystal Tolley for providing access to some specimens used in the study. Johan Marais, Bill Branch, Werner Conradie, Trip Lamb, Don Broadley, Randy Babb, Paul Moler, Ross Sadlier, Glenn Shea, Jens Vindum, and many Villanova University Masters students participated in collecting.
Funding
This project was funded by the University of Michigan (MPH), NSF DEB-0515909 (AMB, TRJ), DEB-0844523 (AMB, TRJ), DEB-1019443 (AMB), DEB-1556255 (AMB), DEB-1556585 (MPH), and EF-1241885 (subaward 13–0632) (AMB). Funding bodies played no role in study design or implementation.
Availability of data and material
The data sets supporting the results of this article are available in the GenBank repository, [accession numbers KY224166–KY224347 in http://www.ncbi.nlm.nih.gov/genbank/].
Authors’ contributions
MPH and AMB designed the project. Data were collected by MPH, TRJ, and AMB. Data analysis was performed by MPH. MPH wrote the paper with input from TRJ and AMB. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Use of animals to obtain tissue samples for genetic analysis was reviewed and approved by the Villanova University Institutional Animal Care and Use Committee. Field work in southern Africa that generated the tissues used in this work was made possible by permits (1987–2014) to AMB from the relevant wildlife authorities of South Africa, Namibia and Zimbabwe.
Abbreviations
- AICc
Corrected Akaike information criterion
- AOO
Area of occupancy
- BI
Bayesian inference
- BiSSE
Binary-state speciation and extinction
- BM
Brownian motion
- bp
Base pairs
- EOO
Extent of occurrence
- GTR
General time reversible model
- HKY
Hasegawa-Kishino-Yano model
- IUCN
International union for the conservation of nature
- K80
Kimura 1980 model
- KIF24
Kinesin family member 24
- LTT
Lineages through time
- Ma
Million years ago
- MCMC
Markov chain Monte Carlo
- ML
Maximum likelihood
- MRCA
Most recent common ancestor
- MuSSE
Multiple-state speciation and extinction
- ND2
NADH dehydrogenase subunit 2
- OU
Ornstein-Uhlenbeck
- PCR
Polymerase chain reaction
- PDC
Phosducin
- RAG1
Recombination activating gene 1
- SH
Shimodaira-Hasegawa
- SVL
Snout-vent length
- TrN
Tamura-Nei model
- tRNA
Transfer RNA
Contributor Information
Matthew P. Heinicke, Email: heinicke@umich.edu
Todd R. Jackman, Email: todd.jackman@villanova.edu
Aaron M. Bauer, Email: aaron.bauer@villanova.edu
References
- 1.IUCN. The IUCN Red List of Threatened Species. Version 2015–4. http://www.iucnredlist.org. Accessed 20 Mar 2016.
- 2.Froese R, Pauly D. FishBase, version (10/2015). http://www.fishbase.org. Accessed 20 Mar 2016.
- 3.Hedges, SB. CaribHerp: West Indian Amphibians and Reptiles. http://www.caribherp.org. Accessed 20 Mar 2016.
- 4.Lamb T, Bauer AM. Footprints in the sand: independent reduction of subdigital lamellae in the Namib–Kalahari burrowing geckos. Proc R Soc Lond B Biol Sci. 2006;273:855–64. doi: 10.1098/rspb.2005.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamble T, Greenbaum E, Jackman TR, Russell AP, Bauer AM. Repeated origin and loss of adhesive toepads in geckos. PLoS One. 2012;7:e39429. doi: 10.1371/journal.pone.0039429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer AM, Branch WR. The herpetofauna of the Richtersveld national park and the adjacent northern Richtersveld, Northern cape Province, Republic of South Africa. Herpetological Nat Hist. 2001;8:111–60. [Google Scholar]
- 7.Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- 8.Losos JB, Mahler DL. Adaptive radiation: the interaction of ecological opportunity, adaptation, and speciation. In: Bell MA, Futuyma DJ, Eanes WF, Levinton JS, editors. Evolution since Darwin: the first 150 years. Sunderland: Sinauer; 2010. pp. 381–420. [Google Scholar]
- 9.Alfaro ME, Brock CD, Banbury BL, Wainwright PC. Does evolutionary innovation in pharyngeal jaws lead to rapid lineage diversification in labrid fishes? BMC Evol Biol. 2009;9:1. doi: 10.1186/1471-2148-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutschmann S, Matschiner M, Damerau M, Muschick M, Lehmann MF, Hanel R, Salzburger W. Parallel ecological diversification in Antarctic notothenioid fishes as evidence for adaptive radiation. Mol Ecol. 2011;20:4707–21. doi: 10.1111/j.1365-294X.2011.05279.x. [DOI] [PubMed] [Google Scholar]
- 11.Deutsch JC. Colour diversification in Malawi cichlids: evidence for adaptation, reinforcement or sexual selection? Biol J Linn Soc. 1997;62:1–4. doi: 10.1111/j.1095-8312.1997.tb01614.x. [DOI] [Google Scholar]
- 12.Masta SE, Maddison WP. Sexual selection driving diversification in jumping spiders. Proc Natl Acad Sci. 2002;99:4442–7. doi: 10.1073/pnas.072493099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiens JJ, Graham CH, Moen DS, Smith SA, Reeder TW. Evolutionary and ecological causes of the latitudinal diversity gradient in hylid frogs: treefrog trees unearth the roots of high tropical diversity. Am Nat. 2006;168:579–96. doi: 10.1086/507882. [DOI] [PubMed] [Google Scholar]
- 14.Bauer AM, Lamb T. Phylogenetic relationships of southern African geckos in the Pachydactylus group (Squamata: Gekkonidae) Afr J Herpetol. 2005;54:105–29. doi: 10.1080/21564574.2005.9635525. [DOI] [Google Scholar]
- 15.Bauer AM, Heinicke MP, Jackman TR, Branch WR. Systematics of the Pachydactylus mariquensis group of geckos (Reptilia: Squamata: Gekkonidae): Status of P. mariquensis latirostris, P. m. macrolepis and P. amoenus. Navorsinge van die Nasionale Museum Bloemfontein. 2011;27:85–107. [Google Scholar]
- 16.Arnold EN. Historical changes in the ecology and behaviour of semaphore geckos (Pristurus, Gekkonidae) and their relatives. J Zool. 1993;229:353–84. doi: 10.1111/j.1469-7998.1993.tb02642.x. [DOI] [Google Scholar]
- 17.Ikeuchi I, Mori A, Hasegawa M. Natural history of Phelsuma madagascariensis kochi from a dry forest in Madagascar. Amphibia-Reptilia. 2005;26:475–83. doi: 10.1163/156853805774806142. [DOI] [Google Scholar]
- 18.Regalado R. Does dichromatism variation affect sex recognition in dwarf geckos? Ethol Ecol Evol. 2015;27:56–73. doi: 10.1080/03949370.2014.885465. [DOI] [Google Scholar]
- 19.Coyne JA. Genetics and speciation. Nature. 1992;355:511–5. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 20.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates; 2004. [Google Scholar]
- 21.Jablonski D. Heritability at the species level: analysis of geographic ranges of Cretaceous mollusks. Science. 1987;238:360–3. doi: 10.1126/science.238.4825.360. [DOI] [PubMed] [Google Scholar]
- 22.Gaston KJ. Species-range size distributions: products of speciation, extinction and transformation. Philos Transact A Math Phys Eng Sci. 1998;353:219–30. [Google Scholar]
- 23.Webb TJ, Gaston KJ. On the heritability of geographic range sizes. Am Nat. 2003;161:553–66. doi: 10.1086/368296. [DOI] [PubMed] [Google Scholar]
- 24.Waldron A. Null models of geographic range size evolution reaffirm its heritability. Am Nat. 2007;170:221–31. doi: 10.1086/518963. [DOI] [PubMed] [Google Scholar]
- 25.Vamosi SM, Vamosi JC. Perspective: Causes and consequences of range size variation: the influence of traits, speciation, and extinction. Front Biogeography. 2012;4:168–177. [Google Scholar]
- 26.Lee MS, Skinner A, Camacho A. The relationship between limb reduction, body elongation and geographical range in lizards (Lerista, Scincidae) J Biogeogr. 2013;40:1290–7. doi: 10.1111/jbi.12094. [DOI] [Google Scholar]
- 27.Slatyer RA, Hirst M, Sexton JP. Niche breadth predicts geographical range size: a general ecological pattern. Ecol Lett. 2013;16:1104–14. doi: 10.1111/ele.12140. [DOI] [PubMed] [Google Scholar]
- 28.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56:701–10. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 29.Branch WR. Field guide to snakes and other reptiles of southern Africa. 3. Struik: Cape Town; 1998. [Google Scholar]
- 30.Gaston KJ, Blackburn TM. Range size-body size relationships: evidence of scale dependence. Oikos. 1996;479–85.
- 31.Lancaster N. Late Quaternary paleoenvironments in the southwestern Kalahari. Palaeogeogr Palaeoclimatol Palaeoecol. 1989;70:367–76. doi: 10.1016/0031-0182(89)90114-4. [DOI] [Google Scholar]
- 32.Partridge TC. The evidence for Cainozoic aridification in southern Africa. Quat Int. 1993;17:105–10. doi: 10.1016/1040-6182(93)90087-V. [DOI] [Google Scholar]
- 33.Thomas DS, Shaw PA. The evolution and characteristics of the Kalahari, southern Africa. J Arid Environ. 1993;25:97–108. doi: 10.1006/jare.1993.1045. [DOI] [Google Scholar]
- 34.Bauer AM. Evolutionary scenarios in the Pachydactylus group geckos of southern Africa: new hypotheses. Afr J Herpetol. 1999;48:53–62. doi: 10.1080/21564574.1999.9651072. [DOI] [Google Scholar]
- 35.Lamb T, Bauer AM. Phylogenetic relationships of the large-bodied members of the African lizard genus Pachydactylus (Reptilia: Gekkonidae) Copeia. 2002;2002:586–96. doi: 10.1643/0045-8511(2002)002[0586:PROTLB]2.0.CO;2. [DOI] [Google Scholar]
- 36.Bauer AM. Phylogeny and biogeography of the geckos of southern Africa and the islands of the western Indian Ocean: a preliminary analysis. In: Peters G, Hutterer R, editors. Vertebrates in the Tropics. Bonn: Zoologisches Forschungsinstitut und Museum A. Koenig; 1990. pp. 274–284. [Google Scholar]
- 37.Heinicke MP, Greenbaum E, Jackman TR, Bauer AM. Phylogeny of a trans‐Wallacean radiation (Squamata, Gekkonidae, Gehyra) supports a single early colonization of Australia. Zool Scr. 2011;40:584–602. doi: 10.1111/j.1463-6409.2011.00495.x. [DOI] [Google Scholar]
- 38.Heinicke MP, Greenbaum E, Jackman TR, Bauer AM. Evolution of gliding in Southeast Asian geckos and other vertebrates is temporally congruent with dipterocarp forest development. Biol Lett. 2012;8:994–997. doi: 10.1098/rsbl.2012.0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinicke MP, Daza JD, Greenbaum E, Jackman TR, Bauer AM. Phylogeny, taxonomy and biogeography of a circum-Indian Ocean clade of leaf-toed geckos (Reptilia: Gekkota), with a description of two new genera. Syst Biodivers. 2014;12:23–42. doi: 10.1080/14772000.2013.877999. [DOI] [Google Scholar]
- 40.Portik DM, Bauer AM, Jackman TR. The phylogenetic affinities of Trachylepis sulcata nigra and the intraspecific evolution of coastal melanism in the western rock skink. Afr Zool. 2010;45:147–59. doi: 10.3377/004.045.0217. [DOI] [Google Scholar]
- 41.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 42.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Laslett D, Canbäck B. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–5. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 45.Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–20. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 49.Reisz RR, Müller J. Molecular timescales and the fossil record: a paleontological perspective. TRENDS Genet. 2004;20:237–41. doi: 10.1016/j.tig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Kluge AG. Cladistic relationships of sphaerodactyl lizards. Am Mus Novit. 1995;3139:1–23. [Google Scholar]
- 51.Iturralde-Vinent MA, MacPhee RD. Age and paleogeographical origin of Dominican amber. Science. 1996;273:1850. doi: 10.1126/science.273.5283.1850. [DOI] [Google Scholar]
- 52.Lee MS, Hutchinson MN, Worthy TH, Archer M, Tennyson AJ, Worthy JP, Scofield RP. Miocene skinks and geckos reveal long-term conservatism of New Zealand’s lizard fauna. Biol Lett. 2009;5:833–7. doi: 10.1098/rsbl.2009.0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutchinson MN. The first fossil pygopodid (Squamata, Gekkota) and a review of mandibular variation in living species. Memoirs Queensland Museum. 1997;41:355–366. [Google Scholar]
- 54.Lee MS, Oliver PM, Hutchinson MN. Phylogenetic uncertainty and molecular clock calibrations: a case study of legless lizards (Pygopodidae, Gekkota) Mol Phylogenet Evol. 2009;50:661–6. doi: 10.1016/j.ympev.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 55.Macey JR, Wang Y, Ananjeva NB, Larson A, Papenfuss TJ. Vicariant patterns of fragmentation among gekkonid lizards of the genus Teratoscincus produced by the Indian collision: a molecular phylogenetic perspective and an area cladogram for Central Asia. Mol Phylogenet Evol. 1999;12:320–32. doi: 10.1006/mpev.1999.0641. [DOI] [PubMed] [Google Scholar]
- 56.Loveridge A. Revision of the African lizards of the family Gekkonidae. Bull Mus Comp Zool. 1947;98:1–469. [Google Scholar]
- 57.Alexander GJ, Marais J. A guide to the reptiles of southern Africa. Cape Town: Struik; 2007. [Google Scholar]
- 58.Bates MF, Branch WR, Bauer AM, Burger M, Marais J, Alexander GJ, De Villiers MS, editors. Atlas and red list of the reptiles of South Africa, Lesotho and Swaziland. Pretoria: South African National Biodiversity Institute; 2014. [Google Scholar]
- 59.Revell LJ. Phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 60.R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 61.Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–45. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 62.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–84. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 63.Pybus OG, Harvey PH. Testing macro–evolutionary models using incomplete molecular phylogenies. Proc R Soc Lond B Biol Sci. 2000;267:2267–72. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beaulieu JM, Jhwueng DC, Boettiger C, O’Meara BC. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution. 2012;66:2369–83. doi: 10.1111/j.1558-5646.2012.01619.x. [DOI] [PubMed] [Google Scholar]
- 65.Collar DC, Schulte II, James A, Losos JB. Evolution of extreme body size disparity in monitor lizards (Varanus) Evolution. 2011;65:2664–80. doi: 10.1111/j.1558-5646.2011.01335.x. [DOI] [PubMed] [Google Scholar]
- 66.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52:131–58. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 67.FitzJohn RG. Quantitative traits and diversification. Syst Biol. 2010;59:619–633. doi: 10.1093/sysbio/syq053. [DOI] [PubMed] [Google Scholar]
- 68.FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–92. doi: 10.1111/j.2041-210X.2012.00234.x. [DOI] [Google Scholar]
- 69.Rabosky DL, Goldberg EE. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst Biol. 2015;64:340–55. doi: 10.1093/sysbio/syu131. [DOI] [PubMed] [Google Scholar]
- 70.Rabosky DL, Huang H. A robust semi-parametric test for detecting trait-dependent diversification. Syst Biol. 2016;65:181–93. doi: 10.1093/sysbio/syv066. [DOI] [PubMed] [Google Scholar]
- 71.Davis MP, Midford PE, Maddison W. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol Biol. 2013;13:1. doi: 10.1186/1471-2148-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bauer AM, Lamb T. Phylogenetic relationships among members of the Pachydactylus capensis group of southern African geckos. Afr Zool. 2002;37:209–20. [Google Scholar]
- 73.Bauer AM, Lamb T, Branch WR. A revision of the Pachydactylus serval and P. weberi groups (Reptilia: Gekkota: Gekkonidae) of Southern Africa, and with the description of eight new species. Proc Calif Acad Sci. 2006;57:595–709. [Google Scholar]
- 74.Lamb T, Bauer AM. Relationships of the Pachydactylus rugosus group of geckos (Reptilia: Squamata: Gekkonidae) Afr Zool. 2000;35:55–67. [Google Scholar]
- 75.Wood PL, Heinicke MP, Jackman TR, Bauer AM. Phylogeny of bent-toed geckos (Cyrtodactylus) reveals a west to east pattern of diversification. Mol Phylogenet Evol. 2012;65:992–1003. doi: 10.1016/j.ympev.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 76.Böhning‐Gaese K, Caprano T, van Ewijk K, Veith M. Range size: disentangling current traits and phylogenetic and biogeographic factors. Am Nat. 2006;167:555–67. doi: 10.1086/501078. [DOI] [PubMed] [Google Scholar]
- 77.Lester SE, Ruttenberg BI, Gaines SD, Kinlan BP. The relationship between dispersal ability and geographic range size. Ecol Lett. 2007;10:745–58. doi: 10.1111/j.1461-0248.2007.01070.x. [DOI] [PubMed] [Google Scholar]
- 78.Olifiers N, Vieira MV, Grelle CE. Geographic range and body size in Neotropical marsupials. Glob Ecol Biogeogr. 2004;13:439–44. doi: 10.1111/j.1466-822X.2004.00115.x. [DOI] [Google Scholar]
- 79.Machac A, Zrzavý J, Storch D. Range size heritability in Carnivora is driven by geographic constraints. Am Nat. 2011;177:767–79. doi: 10.1086/659952. [DOI] [PubMed] [Google Scholar]
- 80.Sinervo B, Calsbeek R, Comendant T, Both C, Adamopoulou C, Clobert J. Genetic and maternal determinants of effective dispersal: the effect of sire genotype and size at birth in side‐blotched lizards. Am Nat. 2006;168:88–99. doi: 10.1086/505765. [DOI] [PubMed] [Google Scholar]
- 81.Beier P, Noss RF. Do habitat corridors provide connectivity? Conserv Biol. 1998;12:1241–52. doi: 10.1111/j.1523-1739.1998.98036.x. [DOI] [Google Scholar]
- 82.Rabosky DL, Lovette IJ. Density-dependent diversification in North American wood warblers. Proc R Soc Lond B Biol Sci. 2008;275:2363–71. doi: 10.1098/rspb.2008.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gavrilets S, Losos JB. Adaptive radiation: contrasting theory with data. Science. 2009;323:732–7. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- 84.Burbrink FT, Pyron RA. How does ecological opportunity influence rates of speciation, extinction, and morphological diversification in New World ratsnakes (tribe Lampropeltini)? Evolution. 2010;64:934–43. doi: 10.1111/j.1558-5646.2009.00888.x. [DOI] [PubMed] [Google Scholar]
- 85.Mahler DL, Revell LJ, Glor RE, Losos JB. Ecological opportunity and the rate of morphological evolution in the diversification of Greater Antillean anoles. Evolution. 2010;64:2731–45. doi: 10.1111/j.1558-5646.2010.01026.x. [DOI] [PubMed] [Google Scholar]
- 86.Pigot AL, Phillimore AB, Owens IP, Orme CD. The shape and temporal dynamics of phylogenetic trees arising from geographic speciation. Syst Biol. 2010;59:660–673. doi: 10.1093/sysbio/syq058. [DOI] [PubMed] [Google Scholar]
- 87.Moen D, Morlon H. Why does diversification slow down? Trends Ecol Evol. 2014;29:190–7. doi: 10.1016/j.tree.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 88.Partridge TC. Of diamonds, dinosaurs and diastrophism: 150 million years of landscape evolution in southern Africa. S Afr J Geol. 1998;101:167–184. [Google Scholar]
- 89.Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin J, Brunet M. Tectonic uplift and eastern Africa aridification. Science. 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- 90.Cowling RM, Proches S, Partridge TC. Explaining the uniqueness of the Cape flora: incorporating geomorphic evolution as a factor for explaining its diversification. Mol Phylogenet Evol. 2009;51:64–74. doi: 10.1016/j.ympev.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 91.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 92.Liu Z, Pagani M, Zinniker D, DeConto R, Huber M, Brinkhuis H, Shah SR, Leckie RM, Pearson A. Global cooling during the Eocene-Oligocene climate transition. Science. 2009;323:1187–1190. doi: 10.1126/science.1166368. [DOI] [PubMed] [Google Scholar]
- 93.Davis CC, Bell CD, Fritsch PW, Mathews S. Phylogeny of Acridocarpus-Brachylophon (Malpighiaceae): implications for Tertiary tropical floras and Afroasian biogeography. Evolution. 2002;56:2395–2405. doi: 10.1111/j.0014-3820.2002.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 94.Kissling WD, Eiserhardt WL, Baker WJ, Borchsenuis F, Couvreur TLP, Balslev H, Svenning J. Cenozoic imprints on the phylogenetic structure of palm species assemblages worldwide. Proc Natl Acad Sci. 2012;109:7379–7384. doi: 10.1073/pnas.1120467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tolley KA, Townsend TM, Vences M. Large-scale phylogeny of chameleons suggests African origins and Eocene diversification. Proc R Soc B. 2013;280:20130184. doi: 10.1098/rspb.2013.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.JRS Biodiversity Foundation grant. Digitizing southwestern-African herpetological collections. http://jrsbiodiversity.org/grants/university-of-florida/. Accessed 1 Aug 2016.
- 97.Hewitt J. Some new forms of batrachians and reptiles from South Africa. Rec Albany Mus. 1935;4:283–357. [Google Scholar]
- 98.Gaston KJ. Rarity. London: Chapman & Hall; 1994. [Google Scholar]
- 99.Harris G, Pimm SL. Range size and extinction risk in forest birds. Conserv Biol. 2008;22:163–71. doi: 10.1111/j.1523-1739.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- 100.Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64:1816–24. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 101.Kozak KH, Wiens J. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution. 2006;60:2604–21. doi: 10.1111/j.0014-3820.2006.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 102.Kozak KH, Weisrock DW, Larson A. Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon) Proc R Soc Lond B Biol Sci. 2006;273:539–46. doi: 10.1098/rspb.2005.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]


