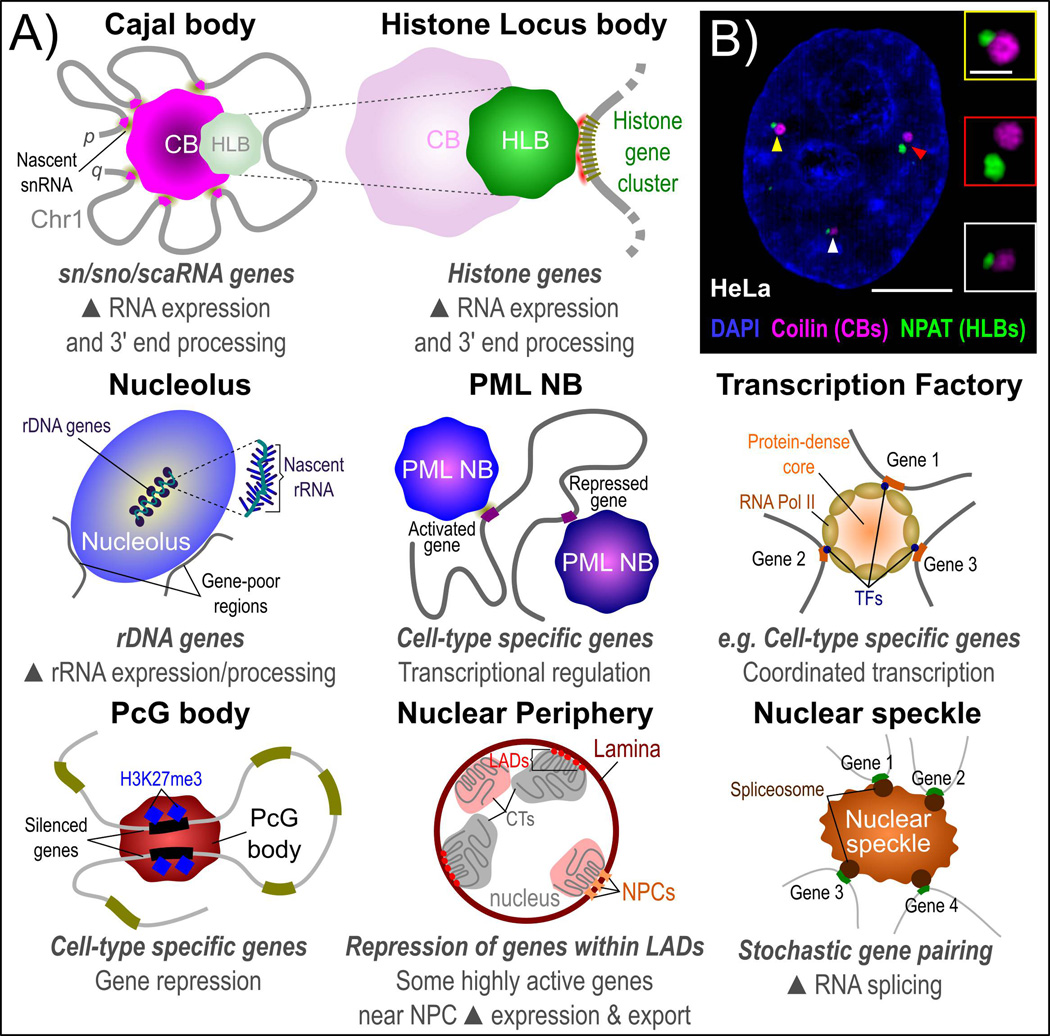
Figure 3.
Nuclear sub-structures and genome organization. A: Many NBs have been reported to form at specific sites of gene activity and influence gene expression. This includes transcriptional activation and repression as well as RNA processing steps. Several NBs have been associated with both gene activation and gene repression, including the role of the nucleolus in activation of ribosomal DNA (rDNA) genes and proximal positioning of gene-poor regions at the nucleolar periphery as well as gene activation and repression by the PML NB, which may be dependent on PML isoform enrichment. Nonrandom physical clustering of specific genes is also reported for non-NB-based structures, such as the nuclear lamina with lamina-associated domains (LADs) and other genes that are highly active and frequently positioned near to the nuclear pore complex (NPC) for efficient export into the cytoplasm. B: Maximally-projected super resolution structured-illumination microscopy (SIM) image of the CB structural protein coilin (far red), the histone transcription factor NPAT (HLBs, green) and DNA (DAPI, blue) in aneuploid HeLa human cervical carcinoma cells. Enlarged insets with positions marked by arrowheads demonstrate frequent physical association between CBs and HLBs. Scale bars represent 5 µm (whole cell) or 1 µm (inset). Images were acquired using a Zeiss Elyra super resolution microscope equipped with a 100× apochromatic 1.46 NA oil immersion objective. 3D-SIM images were generated using Zeiss Zen black software.