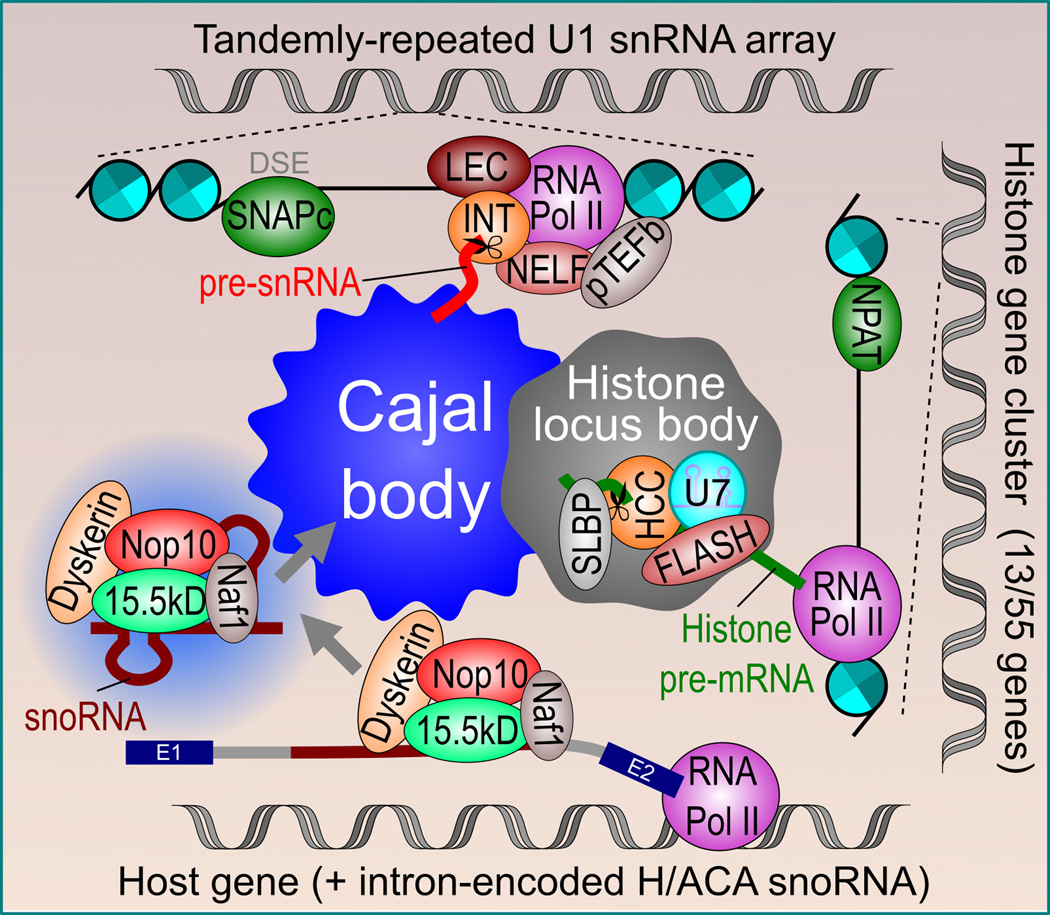
Figure 4.
Cajal body association with target sn/snoRNA and histone genes. CB nucleation and formation is most likely to occur at highly transcriptionally active snRNA gene arrays, such as the tandemly-repeated U1 snRNA gene array on chromosome 1. Transcription of snRNA genes is activated by the binding of the SNAPc transcription factor to the distal sequence element (DSE). Transcription is regulated by the Little Elongation Complex (LEC) and Integrator complex (INT), which is responsible for snRNA 3’ end trimming, as well as accessory factors such as NELF and pTEFb. Intron-encoded snoRNAs are co-transcriptionally isolated from host gene introns by the binding of CB-enriched core components of box H/ACA snoRNAs (e.g. 15.5 kD, Nop10 and dyskerin), which may physically target these regions to the CB periphery. At this point, coilin may bind to excised snoRNPs and help target these nascent snoRNPs to nearby CBs. Finally, through its physical association with the HLB, the major and minor histone gene clusters on chromosomes 6 and 1, respectively, are targeted to the CB periphery. Following transcriptional activation by NPAT, histone pre-mRNAs are processed by U7 snRNP, FLASH and the histone cleavage complex (HCC) within the HLB before binding to the stem-loop binding protein (SLBP).