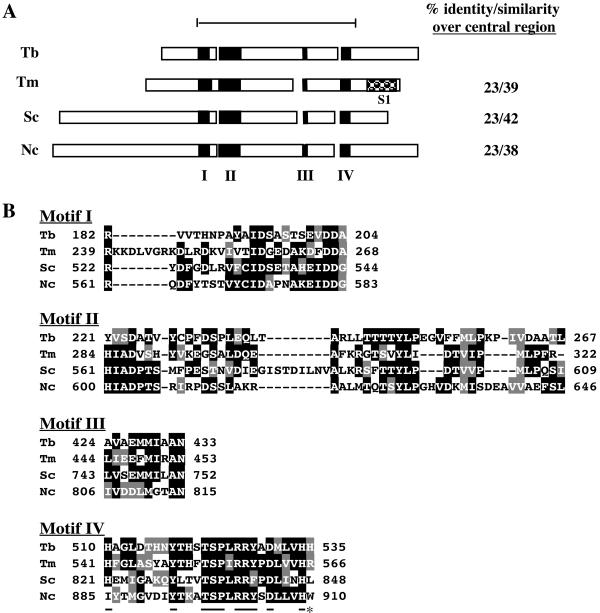
FIG. 1.
Alignment of TbDSS-1 (Tb; accession no. AY233297) with T. maritima RNase R (Tm; accession no. Q9WZI1), S. cerevisiae DSS-1 (Sc; accession no. AAC49144), and Neurospora crassa cyt-4 (Nc; accession no. P47950). (A) Schematic representation of the structure of TbDSS-1 and other RNR family members. Black boxes indicate conserved motifs I to IV. The checked box indicates an S1 RNA binding motif that is present in all RNR family exoribonucleases, with the exception of the mitochondrial members of the family. The bar at the top defines the conserved central region encompassing amino acids 191 to 621 of TbDSS-1, and amino acid conservation in this region is listed on the right. (B) Alignment of motif I to IV sequences. Amino acids that are identical in at least two of the four proteins are indicated by white letters on a black background. Conservative substitutions are indicated by white letters on a gray background. Residues that are underlined in motif IV are highly conserved RNase II signature residues. The C-terminal residue indicated by an asterisk is the highly conserved arginine that is typically absent in mitochondrial RNR family members.