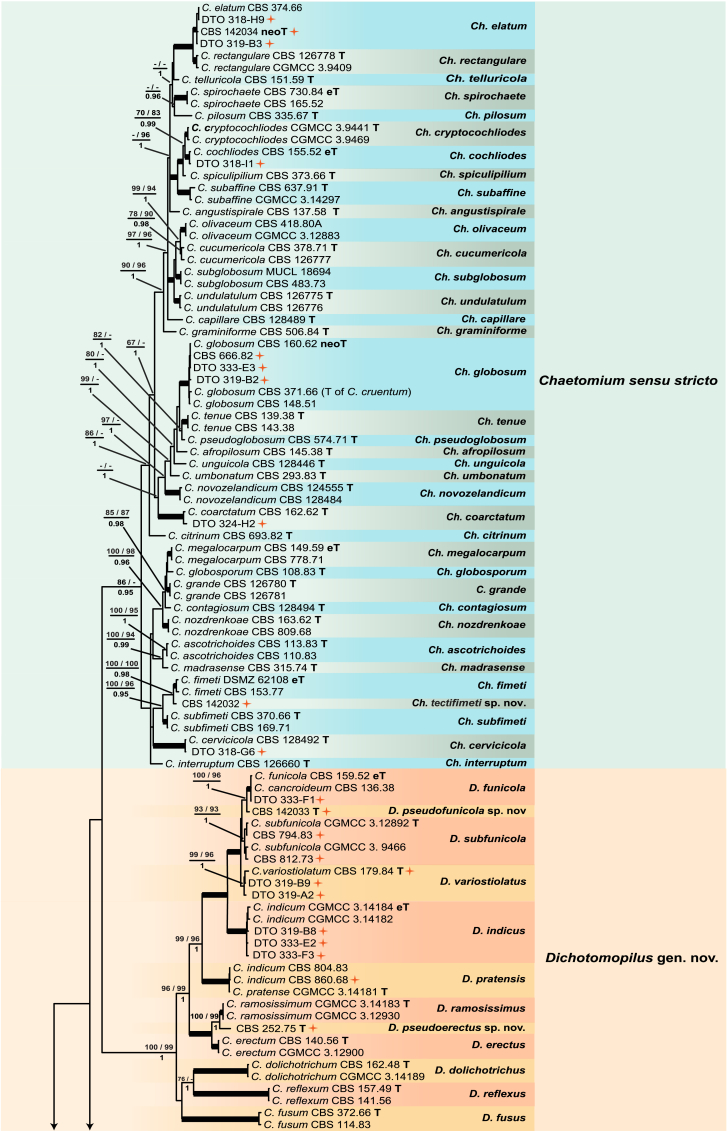
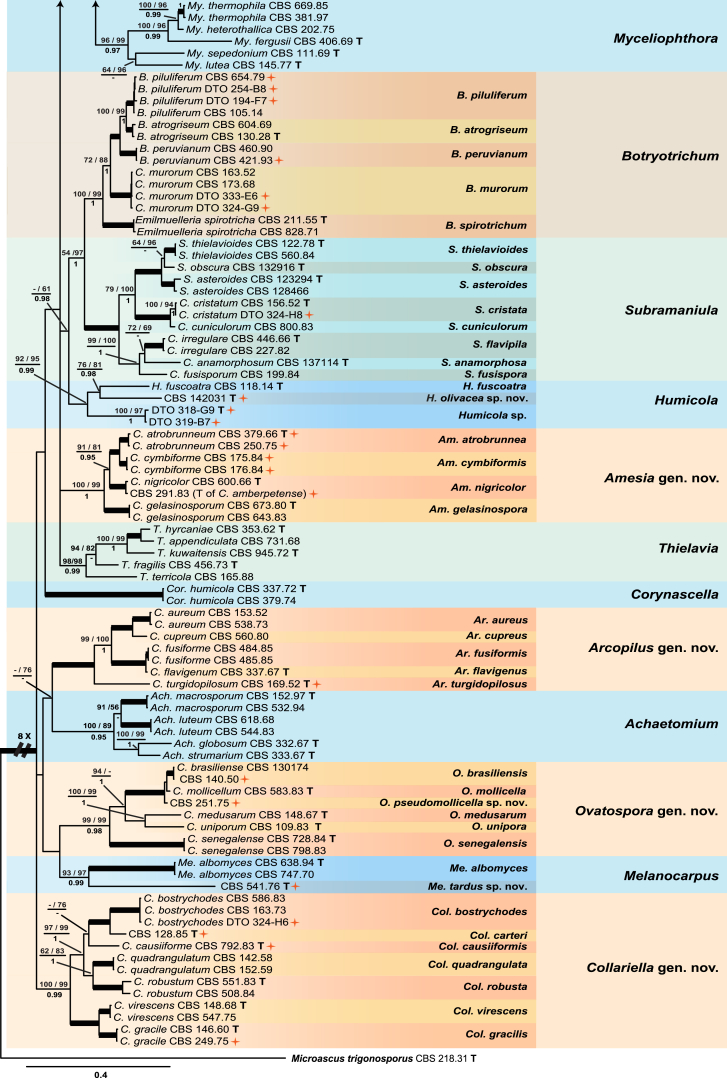
Fig. 1.
Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the concatenated rpb2, tub2, ITS and LSU gene region alignment, with the confidence values of bootstrap proportions from the MP analysis (before the backslash), the ML analysis (after the backslash) above branches, and the posterior probabilities from the Bayesian analysis below branches. The “−” means lacking statistical support (<50 % for bootstrap proportions from ML or MP analyses; <0.95 for posterior probabilities from Bayesian analyses). The branches with full statistical support (MP-BS = 100 %; ML-BS = 100 %; PP = 1.0) are highlighted by thickened branches. Generic novelties are indicated with “gen. nov.” after the genus name and the genus names of the species are abbreviated to facilitate layout of the tree. Genus and species clades are discriminated with boxes of different colours. The 45 isolates from the indoor environment are indicated with a red star on the right side of the culture number; these isolates are representative of all the indoor species recognised in this study. The scale bar shows the expected number of changes per site. The tree is rooted with Microascus trigonosporus strain CBS 218.31 (see Table 1 for GenBank accession numbers).