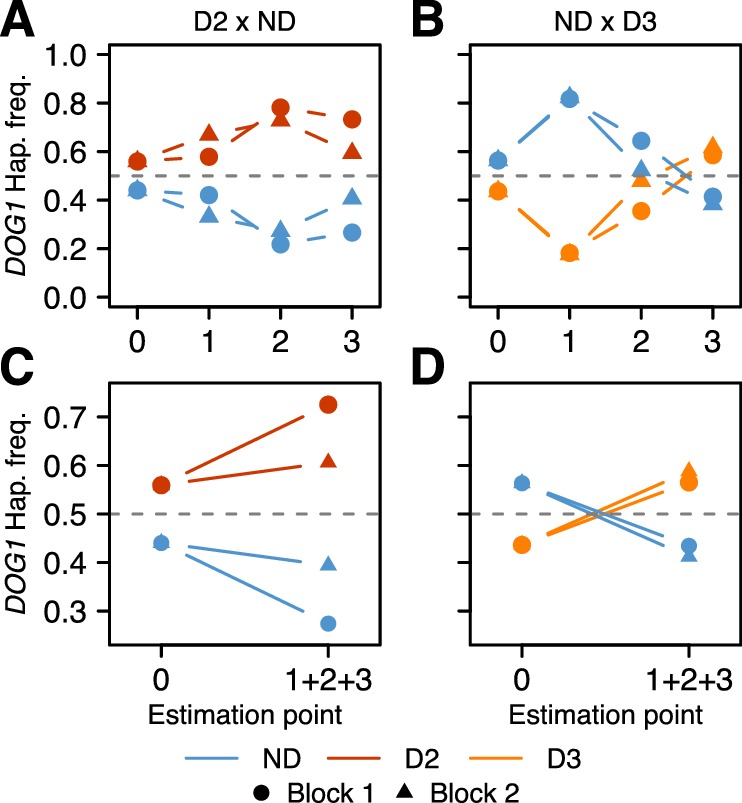
Figure 4. Changes in DOG1 haplotype frequency in southern Sweden field experiments.
(A and C) Frequencies in T880 (D2) x Grön 14 (ND), in which SNP2 segregates. (B and D) Frequencies in Bar 1 (ND) x Ull2-5 (D3), in which SNP3 segregates. (A–B) Frequency changes across time. Starting frequencies were estimated prior to seed dispersal and adjusted to take into account seed viability (0). Frequencies were then estimated in September (1) and November (2) by genotyping seedlings. Finally, seedlings generated from soil samples were used to estimate frequencies in the seed bank (3). (C–D) Difference between starting (0) and ending haplotype frequencies (weighted sum of 1 + 2 + 3).
DOI: http://dx.doi.org/10.7554/eLife.22502.015