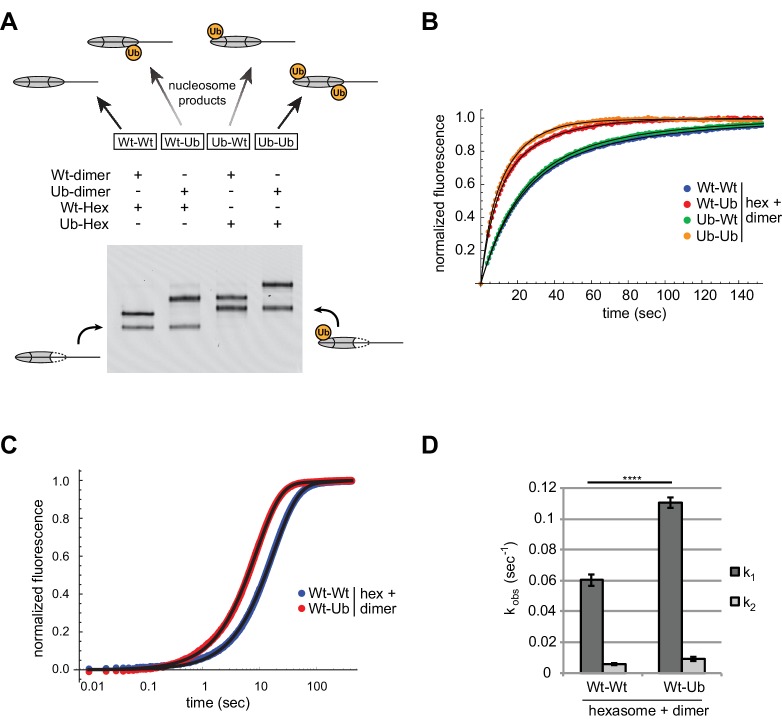
Figure 8. Entry-side H2B-Ubiquitin stimulates nucleosome sliding by Chd1.
(A) Generation of symmetric and asymmetric nucleosomes with site-specific placement of H2B-Ubiquitin. Nucleosomes were formed from subsaturating H2A/H2B dimer (12 nM) addition to 0-601-80 hexasomes (10 nM). Hexasomes and H2A/H2B dimer contained either unmodified (Wt) or ubiquitinated (Ub) H2B as indicated, and resulting nucleosome and hexasome species were visualized by native PAGE. Shown is a representative from six independent dimer addition experiments. (B) Comparison of remodeling reactions with subsaturating (25 nM) Chd1, using hexasomes (10 nM) and H2A/H2B dimers (12 nM) containing unmodified or Ub-conjugated H2B. Shown are progress curves for remodeling reactions monitored using a Cy3-Cy3 pair at 25 μM ATP. Black traces represent fits to the data. Progress curves are representative of two independent experiments. (C) Representative progress curves of nucleosome sliding reactions monitored by stopped flow using Cy3B-Dabcyl at 25 μM ATP and saturating (400 nM) Chd1. Each progress curve is an average of 3–6 technical replicates. Black traces represent fits to the data. (D) Comparison of observed sliding rates monitored with Cy3B-Dabcyl at 25 μM ATP and saturating Chd1 (400 nM). Error bars show standard deviations from three independent experiments. **** p-value <0.0001.