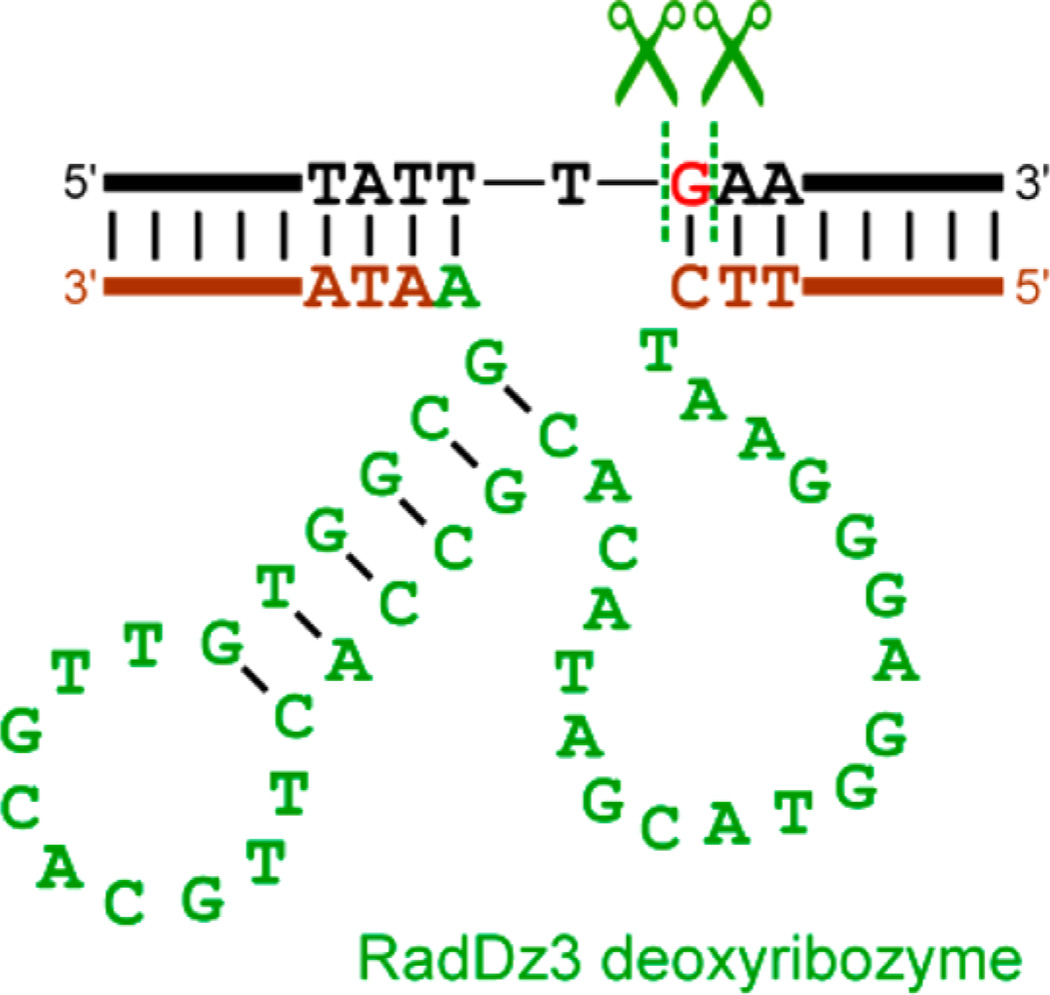
Figure 7.
Mfold-predicted25 secondary structure of the RadDz3 deoxyribozyme (green and brown), shown in association with the DNA substrate that RadDz3 cleaves (black; the excised G nucleoside is red). The RadDz6 deoxyribozyme has numerous alternative predicted secondary structures within 2 kcal/mol, none of which are shown here. See the Experimental Section for full sequences of the fixed segments (brown) that flank the initially random N40 region of RadDz3 (green) as well as the full RadDz6 sequence. The predicted 9 nt loop (TTGCACGTT) of RadDz3 was changed to 4 nt TTTT or GCGA. In each case, the shortened deoxyribozyme retained full activity (data not shown), consistent with but not proving the stem-loop structure. Further experiments to evaluate the RadDz3 secondary structure have not yet been performed.