Figure 1.
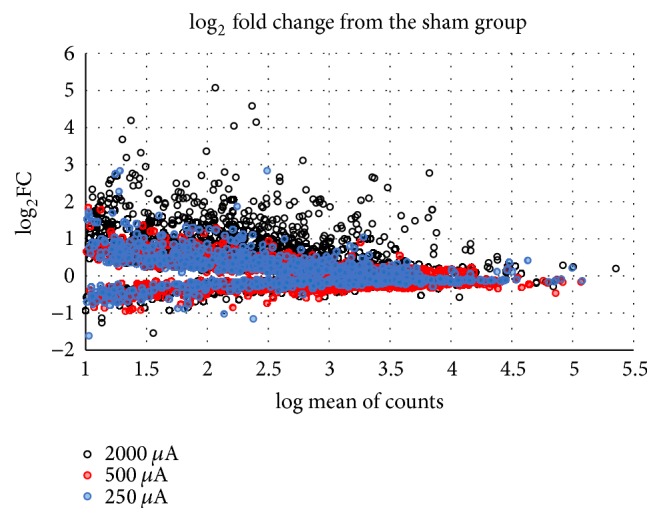
Comparison of genes altered by various tDCS intensities. For this plot, DESeq countable data was used to calculate log10 mean of counts and log2 fold change. The x-axis is log10 mean of counts and the y-axis is log2 fold change from sham group. For this plot, the genes with an expression level ≥ 10 counts and p value (one-tailed t-test between each tDCS and sham group) less than 0.05 were selected to draw the plot. The greater distance from 0 log2FC represents a larger change in gene expression levels from sham.
