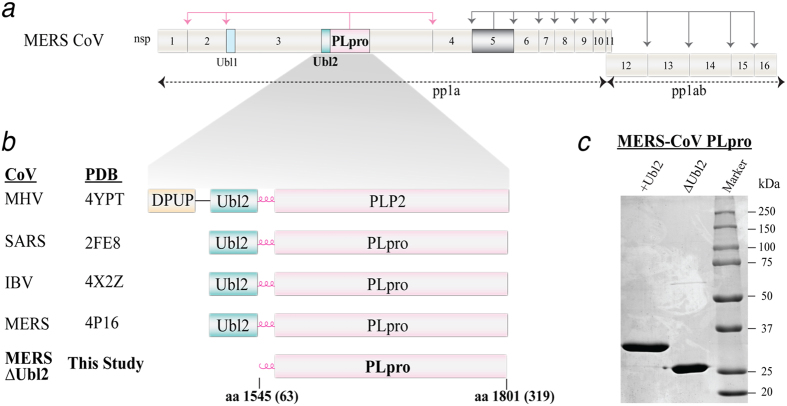
Figure 1. MERS-CoV polyprotein organization and design rationale for the MERS-CoV PLpro-ΔUbl2 construct of nsp3.
(a) Non-structural proteins (nsps) are numbered 1-16 within the MERS-CoV viral polyprotein 1a and 1ab. MERS PLpro is colored in pink in nsp3 and 3CLpro, which is in nsp5, is colored in gray and their respective cleavage sites are colored accordingly and indicated by arrows. The Ubl1 (light blue) and Ubl2 (green) domains of nsp3 are indicated. (b) Summary of the current PLP X-ray structures and the smallest catalytic unit determined in this study. The PDB codes of the X-ray structures that were determined first for MHV PLP2, SARS-CoV, IBV and MERS-CoV PLpro, all containing the Ubl2 domain, are indicated. (c) SDS-PAGE (12.5%) analysis of purified MERS-CoV PLpro-Ubl2 (36 kDa) and −ΔUbl2 (29 kDa) used for activity assays and crystallization. The proteins are estimated to be >95% purity by densitometry.