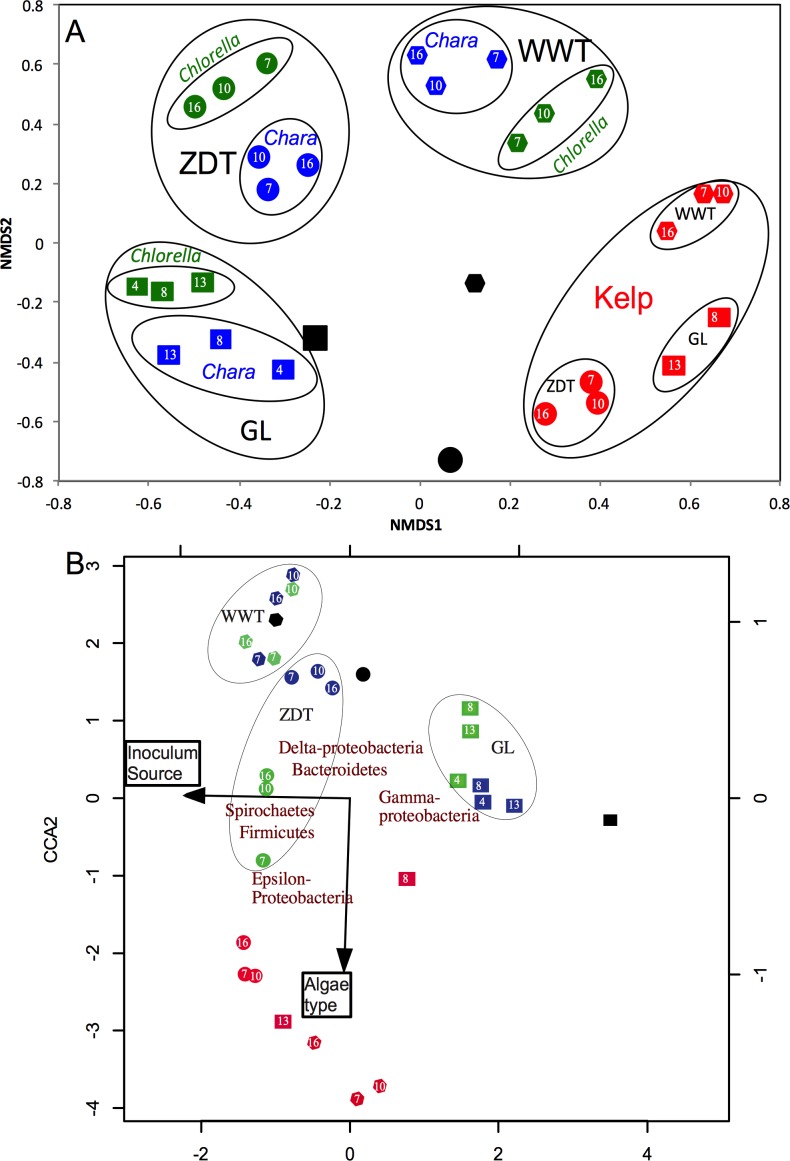
Figure 2. Microbial community structure analysis in the enrichment microcosms (n = 26) as compared to the pre-enrichment inoculum sources (n = 3).
The inoculum sources are denoted by shapes; ZDT (●), WWT (⬣), and GL (■), and the algae types are denoted by color; Chara (blue), Chlorella (green), Kelp (red), and no algae, i.e., pre-enrichment community, (black). Each enrichment condition (inoculum source × algae type) is represented by 3 sample points corresponding to the weeks during enrichment, except for GL-kelp enrichment where the dataset from week 4 is not shown due to the small number of sequences obtained with this dataset. (A) Non-metric multidimensional scaling plots based on Bray–Curtis dissimilarity indices at the species level (0.03). For Chara and Chlorella enrichments, communities grouped by the inoculum source, while Kelp enrichments grouped by the algae type. (B) Canonical correspondence analysis using the abundant phyla/classes relative abundances to study the effect of algae type and inoculum source on the microbial community composition. Here, the same pattern is observed at the phylum/class level, where the community structure of Chara and Chlorella enrichments were similar and grouped by inoculum source, while the microbial community of Kelp enrichments were quite distinct and grouped together regardless of the inoculum source. This pattern is reflected on the direction of the factors arrows, where the algae type is pointing in the direction of the Kelp enrichments. The CCA also depicts the abundant phyla/classes that seem to shape the microbial community in the different enrichments; Gamma-Proteobacteria in GL Chara and Chlorella enrichments, Spirochaetes and Firmicutes in ZDT-Chlorella enrichment, Delta-Proteobacteria and Bacteroidetes in ZDT-Chara enrichments and WWT Chara and Chlorella enrichments, and Epsilon-Proteobacteria and Firmicutes in Kelp enrichments regardless of the inoculum source. The constrained variables explained 57% of the variance.