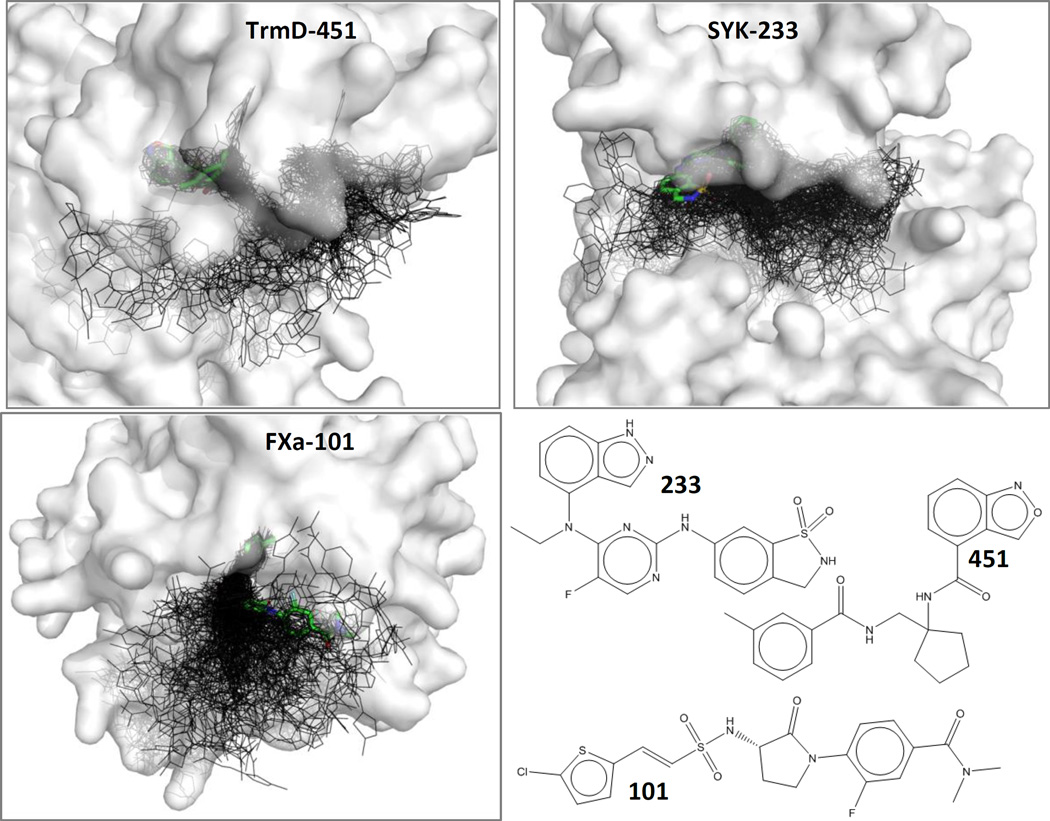
Figure 1.
Examples are given for TrmD, SYK, and FXa, showing the near-native poses (thick sticks with green carbons) among each set of 199 decoys (black lines). Protein surfaces are shown in white and are partially transparent. Ligands are labeled with a short-hand notation above; the complexes are TrmD-gtc000451, SYK-gtc000233, and FXa-gtc000101. These three ligands have the most favorable binding affinity, out of the ligands that have an available crystal structure.