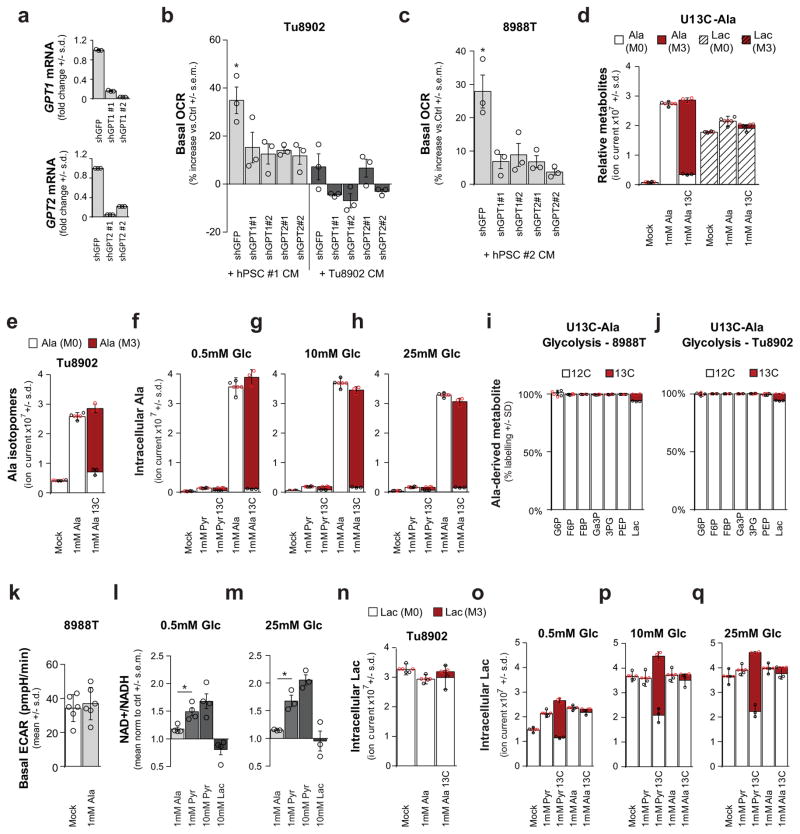
Extended Data Figure 3. Alanine secreted by stellate cells is used by PDAC to fuel biosynthetic reactions.
a–c, Knockdown of GPT1 or GPT2 in PDAC cells (a) significantly attenuates the ability of hPSC-conditioned medium to increase OCR in Tu8902 cells (b). This observation was repeated with conditioned medium from an independent hPSC line in 8988T cells (c). Error bars represent the s.e.m of 3 independent experiments. One-way ANOVA; b, *P =0.0134; c, *P =0.0129. d–j, Metabolic tracing studies using U-13C-Ala and U-13C-pyruvate (Pyr) (f–h). d, Metabolic tracing studies using U-13C-Ala in 8988T cells. Error bars, s.d. of n = 3. Two-way ANOVA was performed: for alanine, ***P < 0.0001; for lactate, ***P =0.0001, **P =0.0086. e, Intracellular accumulation and labelling of alanine in Tu8902 cells treated with 1 mM alanine. Two-way ANOVA was performed: ***P < 0.0001. f–h, Intracellular accumulation and labelling of alanine in 8988T cells treated with 1 mM pyruvate or 1 mM alanine grown in media containing different glucose (Glc) concentrations (0.5, 10, 25 mM). Error bars represent s.d. of n =3. i, j, U-13C-Ala does not contribute to glycolysis or gluconeogenesis as seen for the metabolites glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), fructose bis-phosphate (FBP), glyceraldehyde 3-phosphate (Ga3P), 3-phosphoglycerate (3PG), and phosphoenolpyruvate (PEP) in 8988T (i) or Tu8902 (j) PDAC cell lines. Label can be incorporated in lactate (Lac) independent of glycolysis. Error bars represent s.d. of n =3. k, 1 mM alanine does not significantly increase basal extra cellular acidification rate (ECAR) of 8988T cells. Error bars represent s.d. of 6 replicates. Data presented for a representative experiment (of 3 experiments), P =0.6082. l, m, Alanine does not alter the NAD+/NADH ratio in 8988T cells to the same extent as pyruvate in medium containing either 0.5 mM (l) or 25 mM (m) extracellular glucose. 10 mM pyruvate and 10 mM lactate were included as controls; error bars represent s.e.m of n =4 (l) or n =3 (m) independent experiments. t-test performed; l, *P =0.0205; m, *P =0.0291. n–q, Alanine contributes minimally to lactate in both Tu8902 (n) and 8988T (o–q) cells, and independently of glucose concentrations in medium, as seen by tracing of U-13C-Ala: 0.5 mM (o), 10 mM (p) or 25 mM (q) glucose. Pyruvate labels ~50% of the lactate pool in 8988T cells (o–q) independent of the glucose concentration in the medium. Error bars represent the s.d. of n =3. d–j, n–o, n = 3 technical replicates from independently prepared samples from individual wells