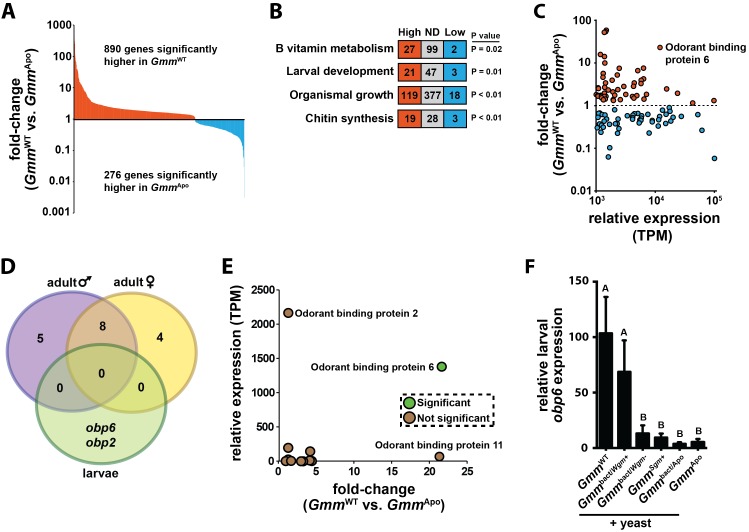
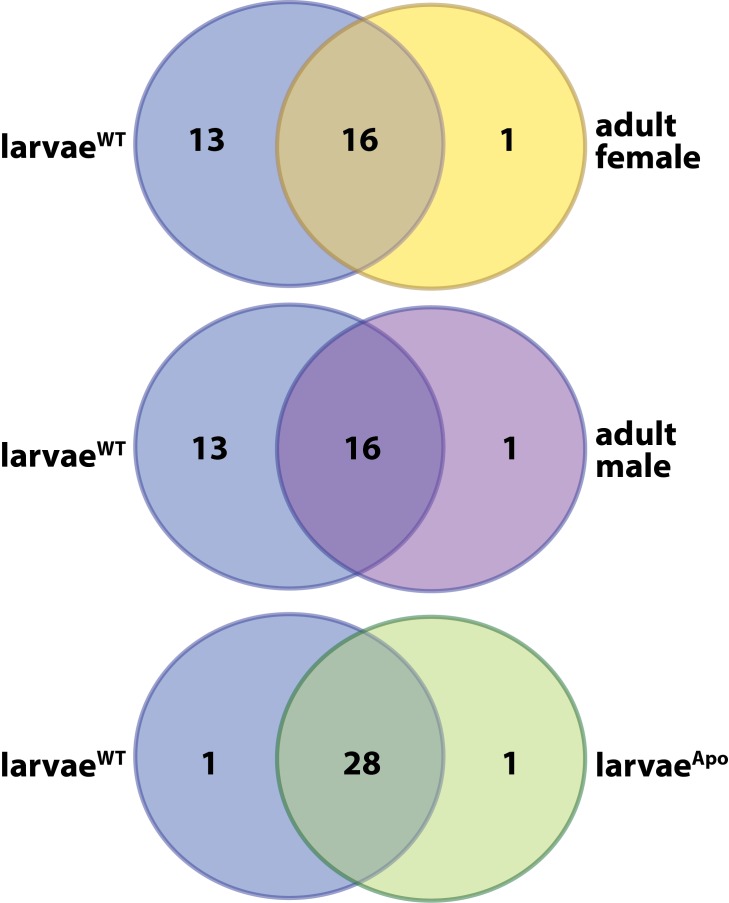
Figure 1. Symbiont-mediated differential expression of odorant binding protein 6 in tsetse larvae.
(A) Number of genes exhibiting significant differential expression, and a relative transcript abundance [in transcripts per million (TPM)] over 3, in GmmWT compared to GmmApo larvae. Significance is based on a Baggerly’s test followed by a false detection rate correction (p<0.01). (B) Significantly enriched gene ontology categories, determined using a Fisher’s exact test. (C) Genes exhibiting significant differential expression (measure as fold-change in gene expression) in GmmWT compared to GmmApo larvae, and a minimum TPM value of 1000. Significance was determined as in (A). (D) Enrichment analysis of odorant binding protein-encoding genes expressed in GmmWT adult males (purple) and females (yellow), and GmmWT larvae (green). (E) Relative expression (TPM) of tsetse odorant binding protein-encoding genes in GmmWT larvae, and their differential expression (measure as fold-change in gene expression) in GmmWT compared to GmmApo larvae. Significance was determined as in (A). (F) Relative obp6 expression in GmmWT larvae, as well as larvae derived from symbiont-cured moms fed a diet supplemented with yeast and Wigglesworthia-containing bacteriome extracts (Gmmbact/Wgm+), Wigglesworthia-free bacteriome extracts (Gmmbact/Wgm-), Sodalis cell extracts (GmmSgm+), and bacteriome extracts harvested from GmmApo females (Gmmbact/Apo). GmmWT and GmmApo flies served as controls. n = 6 biological replicates for groups GmmWT, Gmmbact/Wgm+ and GmmSgm+ samples, and n = 5 biological replicates for Gmmbact/Wgm-, Gmmbact/Apo and GmmApo samples. Replicates for all groups contain a mixture of four first and second instar larvae. Data are presented as mean ± SEM. Bars with different letters indicate a statistically significant difference (specific p values are listed in Figure 1—source data 1) between samples. Statistical analysis = ANOVA followed by Tukey’s HSD post-hoc analysis.
DOI: http://dx.doi.org/10.7554/eLife.19535.003