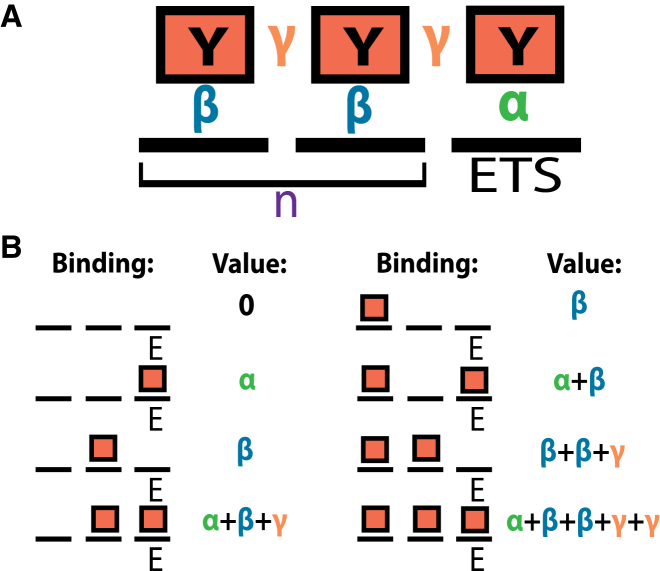
Figure 1.
Lattice model for Yan binding at equilibrium. (A) Yan molecules (boxes) occupy discrete sites (bold black lines) at an element of DNA. The class of elements considered by the model contain one high-affinity specific binding site (ETS site) and n non-specific binding sites. Yan molecules participate in different types of interactions depending on their occupancy at the element, and the free energies of these interactions are given by the terms α, β, and γ. α represents the specific DNA binding free energy, β represents the non-specific DNA binding free energy, and γ represents the SAM-mediated protein-protein interaction free energy, contingent upon two molecules of Yan being adjacent to one another in the lattice. (B) Configurations of binding and free energy values for an element where n is equal to 2. The binary representation of the index of configurations (0,1,…,6,7) corresponds to the binding configuration itself. This provides a straightforward means of manipulating and scoring many microstates. Each microstate is shown with its free energy in terms of α, β, and γ. To see this figure in color, go online.