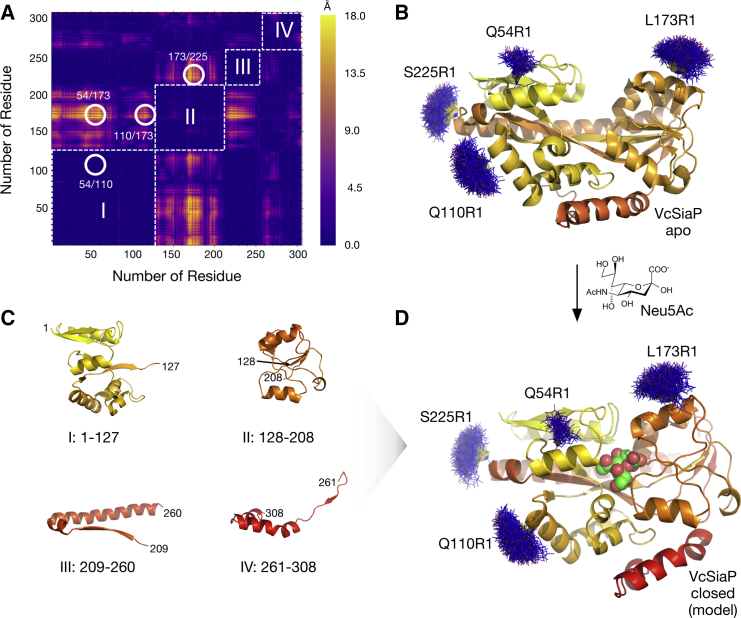
Figure 1.
Structural changes of P domains. (A) Difference distance matrix (diffDM) for the substrate-bound and -free forms of HiSiaP (PDB: 3B50 (37), PDB: 2CEY (10)). (Dark violet regions) Pairs of residues, where the Cβ-Cβ distance does not change between both conformations. (Yellow peaks) Large distance changes of up to 18 Å. (White circles) Pairs of residues that were selected as spin labeling sites. The violet squares along the diagonal of the matrix (dashed, white lines) can be interpreted as rigid domains (I–IV) of the P domain. Note that the matrix is symmetric along its diagonal. (B) The substrate-free structure of VcSiaP (PDB: 4MAG (23)). The protein is shown as cartoon model. A color gradient is running from yellow (N-terminus) to red (C-terminus) to indicate the trace of the polypeptide chain. Models of spin labels at positions highlighted in (A) were attached with MtsslWizard (blue lines). (C) Cartoon models of the individual structures of the rigid domains I-IV of substrate-free VcSiaP. (D) Model of the closed form of VcSiaP. The model was produced by superposing the rigid domains I-IV in (C) onto the structure of closed HiSiaP (PDB: 3B50 (37)). The model of the bound Neu5Ac is shown as spheres.