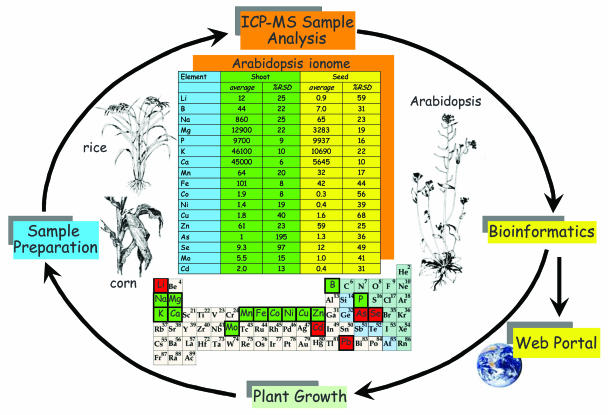
Figure 1.
High-throughput ionomics. Putative mutants and wild-type Arabidopsis plants are grown together with known ionomic mutants, used as positive controls, under standardized conditions. Plants are uniformly sampled, digested in concentrated nitric acid, diluted, and analyzed for numerous elements using ICP-MS. Raw ICP-MS data are normalized using analytical standards and calculated weights based on wild-type plants (Lahner et al., 2003). Data are processed using custom tools and stored in a searchable, World Wide Web-accessible database. Ionomic analysis can also be applied to other plants with available genetic resources, including rice and maize. Elements in the Periodic Table highlighted in black boxes represent those elements analyzed during our ionomic analyses using ICP-MS, elements highlighted in green are essential for plant growth, and those in red represent nonessential trace elements. The table represents Arabidopsis (Col 0) shoot and seed ionomes, all elements presented as μg g−1 dry weight. Data represent the average shoot concentrations from 60 individual plants and seed from 12 individuals ± sd as percentage of average (%RSD), all plants grown as described by Lahner et al. (2003).