Figure 2.
Molecular and Pluripotency Features of Vitamin C- and l-Proline-Induced Cells
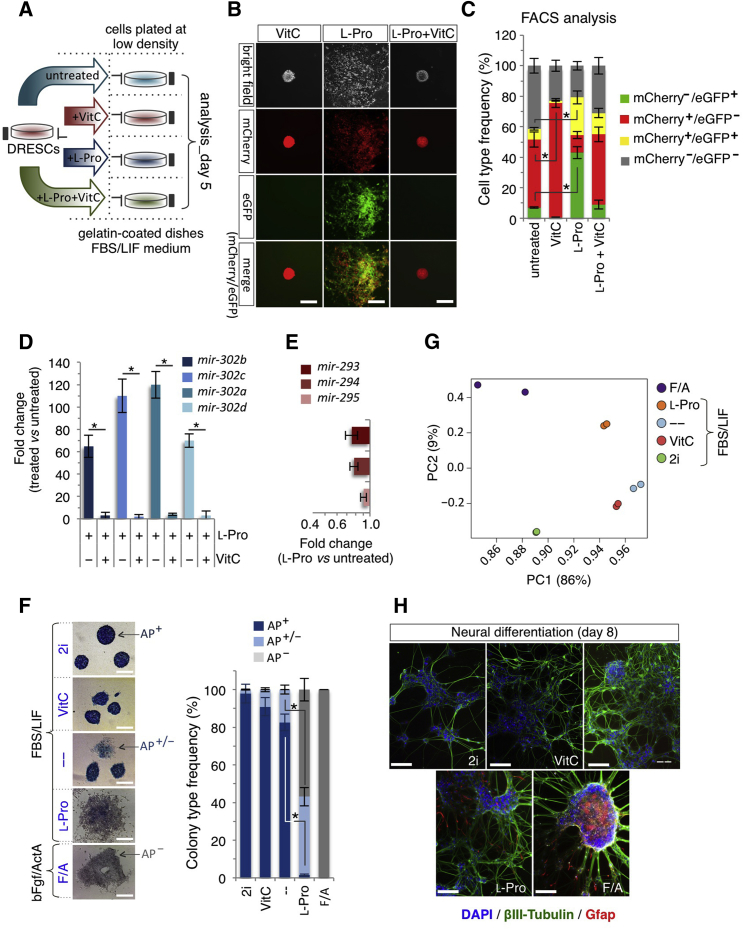
(A) Schematic representation of the experimental strategy. Dual-reporter ESCs (DRESCs) plated at low density ± VitC (500 μM)/± l-Pro (150 μM) and analyzed at day 5.
(B) Representative bright-field and fluorescence images of colonies generated from DRESCs ± VitC (500 μM)/± l-Pro (150 μM) at day 5. Red (mir-290), green (mir-302), and yellow (mir-290 and mir-302) signals indicate expression of the transgenes. Scale bars, 250 μm.
(C) Fluorescence-activated cell sorting (FACS) quantification of mCherry±/eGFP± cells in DRESCs ± VitC (500 μM)/± l-Pro (150 μM). Data are mean ± SEM; ∗p < 0.01 (n = 3 independent experiments).
(D) qPCR analysis of mir-302 expression in l-Pro (150 μM) ± VitC (500 μM)-treated ESCs at day 5. Data are fold change in gene expression compared with control, normalized to Gapdh, and are mean ± SEM; ∗p < 0.005 (n = 3 independent experiments).
(E) qPCR analysis of mir-290 expression in l-Pro-treated ESCs (150 μM) at day 5. Data are fold change versus control, normalized to Gapdh, and are mean ± SEM (n = 3 independent experiments).
(F) Representative pictures (left) and frequency (right) of AP+, AP+/−, and AP− staining on colonies derived from ESCs plated at low density in the presence of the indicated inhibitors, metabolites, and growth factors. Data are mean ± SEM; ∗p < 0.05 (n = 3 independent experiments). Scale bars, 250 μm.
(G) Principal component (PC) analysis of RNA-sequencing data generated from ESCs grown as indicated in (F).
(H) Representative pictures of βIII-tubulin (green) and GFAP (red) immunofluorescence on neurons derived from ESCs grown as in (F). Nuclei are stained with DAPI. Scale bars, 100 μm.
See also Figure S2.