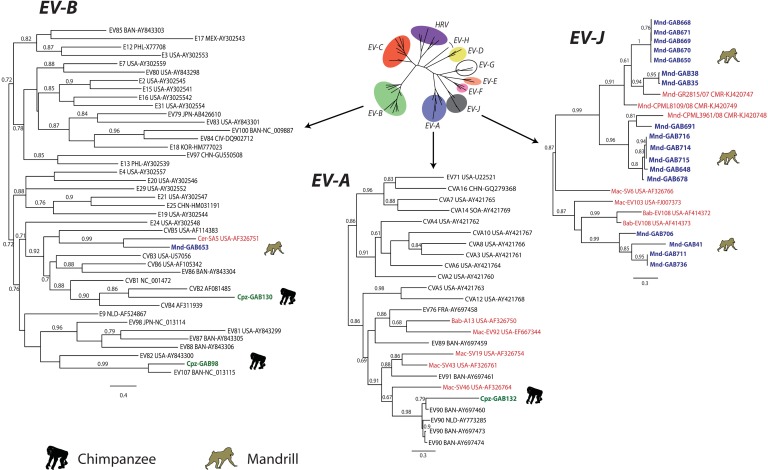
Fig 2. Phylogenetic relationship of EV strains based on approximately 450 nucleotides VP2 sequences (positions 962 to 1,545 according to the poliovirus 1 genome, Genbank # V01149), in the species EV-B, EV-A and EV-J.
The genus tree was constructed using the partial VP2 sequences of representative serotypes of the genus Enterovirus by maximum likelihood method. Sequences obtained in this study are indicated in blue for mandrills and green for chimpanzees. Reference EV sequences of other NHP species are indicated in red. Names of NHP species are indicated (Mac for macaque, Bab for baboon, Cer for cercopothecus), countries are also indicated (BAN, Bangladesh; CHN for China; CIV for Ivory Coast; CMR for Cameroon; FRA for France; JPN for Japan; KOR for South Korea; MEX for Mexico; NLD for Netherland; PHL for Philippines; PUR for Puerto Rico; SLN for Slovenia; USA for United States of America; and SOA for Republic of South Africa). Trees were built using Maximum likelihood algorithm and GTR substitution model. Only bootstrap values ≥ 60% are indicated at the nodes. Scale bars represent the genetic distance. GenBank accession numbers of the sequences used are indicated in the tree (for details, see S4 Table).