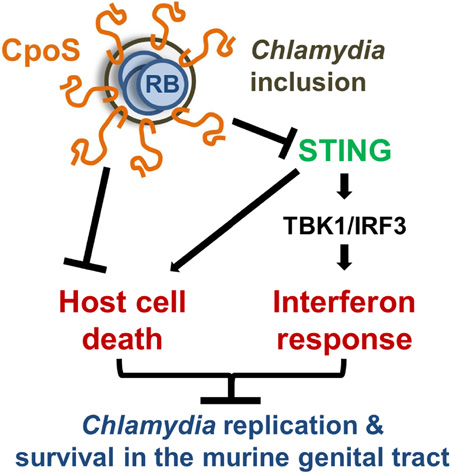
Summary
Evading cell death is critical for Chlamydia to maintain a replicative niche but the underlying mechanisms are unknown. We screened a library of Chlamydia mutants for modulators of cell death. Inactivation of the inclusion membrane protein CpoS (Chlamydia promoter of Survival) induced rapid apoptotic and necrotic death in infected cells. The protection afforded by CpoS is limited to the inclusion in which it resides indicating that it counteracts a spatially restricted pro-death signal. CpoS-deficient Chlamydia induced an exacerbated type I interferon response that required the host cGAS/STING/TBK1/IRF3 signaling pathway. Disruption of STING, but not cGAS or IRF3, attenuated cell death suggesting that STING mediates Chlamydia-induced cell death independent of its role in regulating interferon responses. CpoS-deficient strains are attenuated in their ability to propagate in cell culture and are cleared faster from the murine genital tract, highlighting the importance of CpoS for Chlamydia pathogenesis.
Graphical Abstract
eTOC blurb
Evading cell death is critical for Chlamydia trachomatis to maintain a replicative niche. Sixt et al. show that the Chlamydia effector protein CpoS counteracts STING-mediated pro-death signals and type I IFN responses in response to Chlamydia-containing vacuoles, which enables optimal intracellular bacterial growth and survival in the murine genital tract.
Introduction
The success of intracellular pathogens depends on their ability to evade cell-autonomous defense responses (Randow et al., 2013). For instance, the induction of host cell death can deprive pathogens from a replicative niche (Labbé and Saleh, 2008). This response is especially effective against obligate intracellular pathogens such as Chlamydia trachomatis, a sexually-transmitted pathogen associated with pelvic inflammatory disease and infertility (Mylonas, 2012). Chlamydia spp. alternate between two developmental stages: the non-infectious reticulate body (RB) that replicates within a membrane-bound vacuole (termed inclusion), and the environmentally resistant elementary body (EB) (Ward, 1988). Because the developmental cycle needs to be complete to generate infectious EBs, maintenance of host cell viability during the RB replication phase is critical for the propagation of Chlamydia spp. Candidate Chlamydia effectors that may protect the inclusion include a family of integral inclusion membrane proteins (Incs) (Rockey et al., 2002). In this study, we employed emerging genetic tools in Chlamydia to identify and characterize a C. trachomatis mutant lacking the Inc CpoS. Infection with this mutant resulted in premature death of the host cell, hyper-induction of type I interferons, decreased production of EBs, and rapid clearance from the murine genital tract.
Results
A genetic screen identifies a C. trachomatis mutant with enhanced cytotoxicity
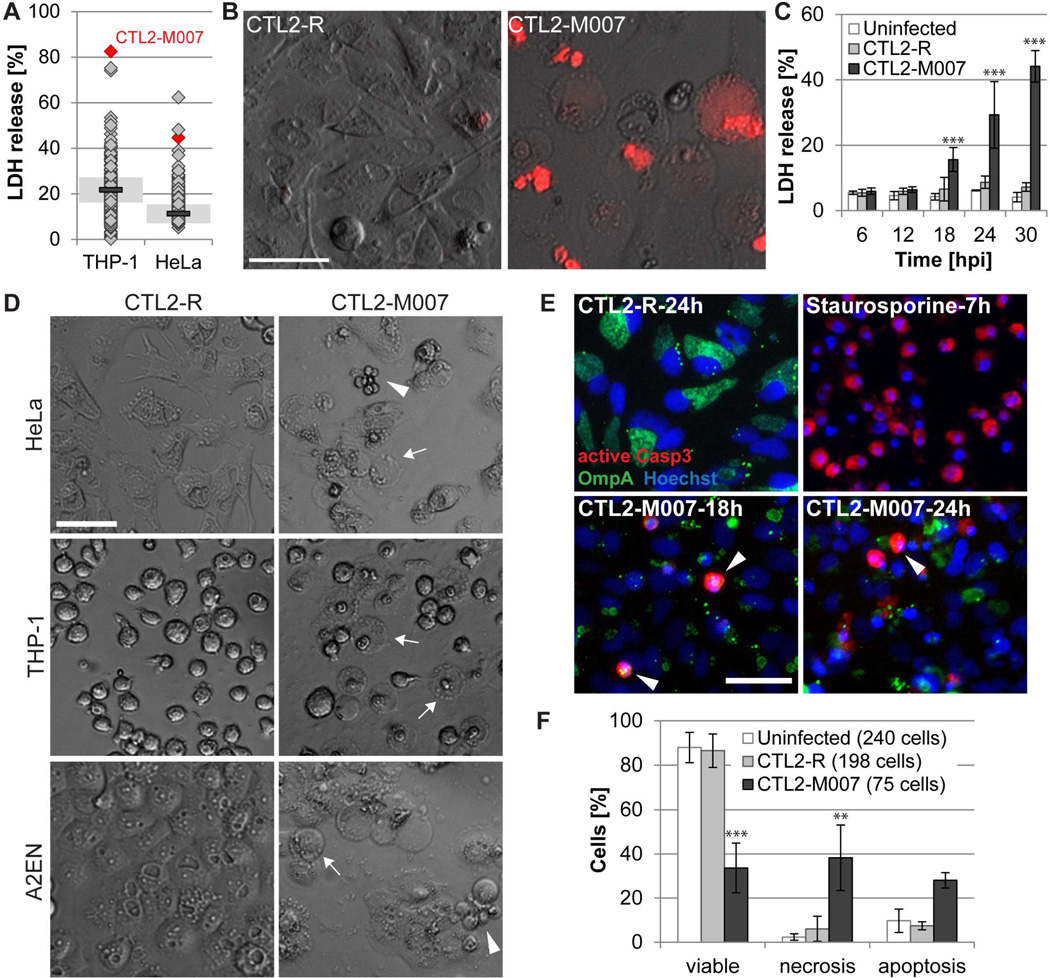
We screened C. trachomatis mutants (Kokes et al., 2015) for strains that induce cytotoxicity in cervical epithelial (HeLa) and monocytic (THP-1) cells at mid-stage of infection. We identified mutant strains that caused the release of higher levels of host lactate dehydrogenase (LDH), as compared to the parent strain CTL2-R (Fig. 1A), and focused our analysis on mutant CTL2-M007, which reproducibly induced enhanced LDH release (Fig. S1A) and host cell permeability to propidium iodide (Fig. 1B). In HeLa cells, death was first detectable at 18 hpi (Fig. 1C). The magnitude of the response was dependent on the bacterial dose (Fig. S1B) and cytotoxicity required bacterial viability and de novo protein synthesis (Fig. S1C). Infections with purified EBs caused similar levels of cell death as crude bacterial preparations (Fig. S1D), indicating that death was not induced by soluble factors.
Figure 1. A genetic screen identifies a C. trachomatis strain that causes apoptotic and necrotic cell death.
(A) Cell lysis induced by Chlamydia mutants as assessed by the release of LDH into supernatants at 28 hpi (THP-1) or 24 hpi (HeLa) (mutants (diamonds, n=224); CTL2-R (bar, mean; shaded area, SD; n=2)).
(B) Loss of membrane integrity during infection of HeLa cells with CTL2-M007 (10 IFU/cell) visualized by enhanced permeability to propidium iodide (red) at 24 hpi (bar=50 µm).
(C) Time course of cell death induction by CTL2-M007 (10 IFU/cell) in HeLa cells (mean±SD, n=3, two-way ANOVA + Newman-Keuls).
(D) Induction of apoptosis (arrowheads) and necrosis (arrows) by CTL2-M007 in epithelial (HeLa, A2EN) and monocytic (THP-1) cells (10 IFU/cell, 21 hpi). Bar=50 µm.
(E) Immunofluorescence detection of apoptotic CTL2-M007-infected (10 IFU/cell) HeLa cells (Chlamydia OmpA, green; active caspase-3, red; Hoechst, blue). Arrowheads: apoptotic infected cells. Bar=50 µm.
(F) Live imaging based assessment of the frequency of apoptosis and necrosis in CTL2-M007-infected (10 IFU/cell) cells until 30 hpi. The category “uninfected” refers to cells in infected wells that contain no inclusions (mean±SD, n=2, two-way ANOVA + Newman-Keuls).
See also Fig. S1 and movies S1–3.
CTL2-M007-infected cells display features of apoptotic and necrotic cell death
CTL2-M007-induced death was also observed in differentiated THP-1 cells, A2EN (human endocervical epithelial) cells, HEK293T (human embryonic kidney) cells, and MEFs (mouse embryonic fibroblasts) (Fig. 1D and S1E), but was less pronounced in Vero cells (Fig. S1F). THP-1 cells died by a necrotic-type of death generating balloon-like cell remnants (Fig. 1D and S1E). In non-phagocytic cells, a proportion of cells adopted an apoptosis-like morphology characterized by cell shrinkage, rounding, and membrane blebbing (Fig. 1D and S1E). Indeed, CTL2-M007-infected HeLa cells displayed hallmarks of caspase-dependent apoptosis, including condensed nuclei, immunoreactivity to antibodies specific for proteolytically mature caspase-3 (Fig. 1E), and caspase-3 activity based on the cleavage of the fluorescent substrate NucView-488 (Fig. S1G) and DEVD cleavage activity in cell lysates (Fig. S1H). These features were similar to those induced by the apoptosis inducer staurosporine (Fig. 1E and S1H). Time-lapse microscopy indicated that by 30 hpi only 34% of the HeLa cells infected with CTL2-M007 remained viable, 28% had died with features of apoptosis, and 38% had disintegrated by necrosis (Fig. 1F; movies S1–3). In sharp contrast, 88% of inclusion-free cells and 87% of cells containing CTL2-R inclusions remained viable during the same time frame.
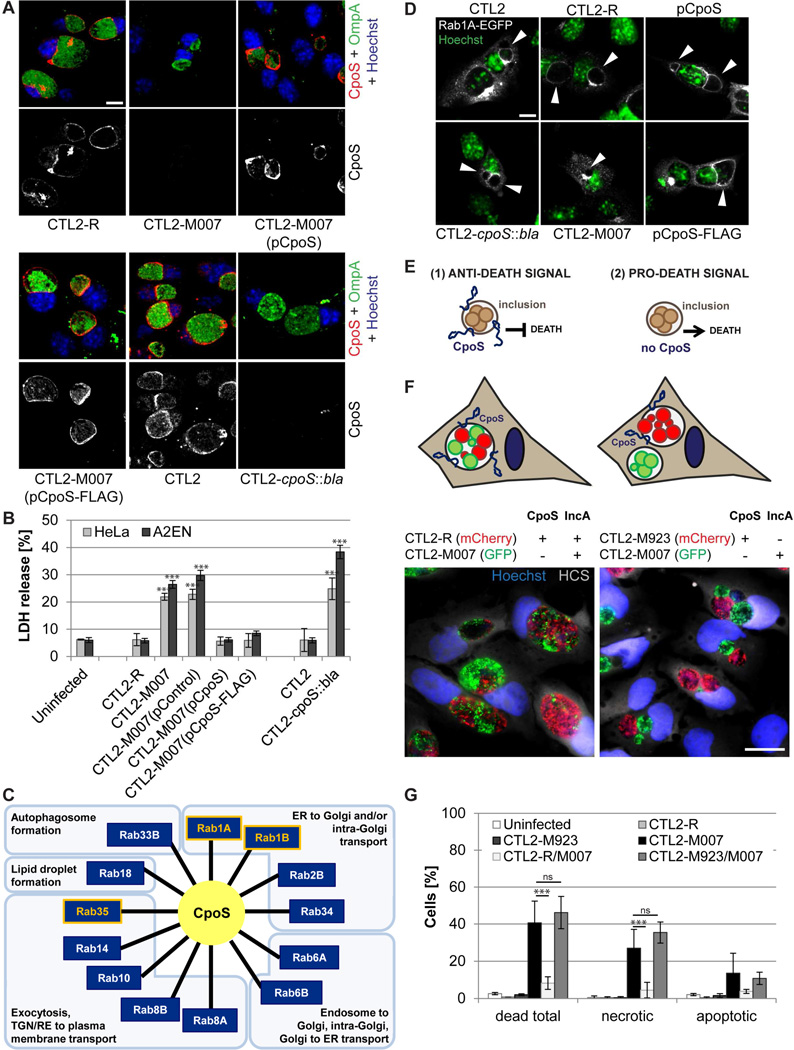
CTL2-M007-induced cytotoxicity is associated with the loss of the Inc CpoS
CTL2-M007 contains 14 mutations (Table S1). We generated recombinants between CTL2-M007 and wild-type bacteria and mapped the mutation responsible for cytotoxicity to a premature stop codon (Q31*) in the gene CTL0481 (CT229 in C. trachomatis serovar D) (Table S2). This gene encodes an Inc protein we refer to as CpoS, for Chlamydia promoter of Survival. We next transformed CTL2-M007 with a plasmid enabling expression of full-length cpoS. In parallel, we disrupted cpoS in CTL2 by insertional gene inactivation with an intron carrying a β-lactamase resistance (bla) cassette (Fig. S2A–B). CpoS was detectable as a 19 kD protein in HeLa cells infected with parental strains but not CTL2-M007 or CTL2-cpoS::bla (Fig. S2C). CpoS expression was significantly increased in cells infected with the complemented mutant (Fig. S2D) and immunofluorescence microscopy indicated that CpoS, as previously shown (Bannantine et al., 2000; Rzomp et al., 2006), localized to the inclusion membrane (Fig. 2A). The cytotoxicity induced by CTL2-M007 was abolished by introduction of the CpoS-expressing plasmid (Fig. 2B). Disruption of cpoS in wild-type bacteria recapitulated the toxicity phenotype observed in CTL2-M007 (Fig. 2B). Overall, these findings provide genetic confirmation that the loss of CpoS induces toxicity in infected cells.
Figure 2. Loss of CpoS on the inclusion causes enhanced death in infected cells.
(A) Immunolocalization of CpoS in HeLa cells infected for 26 h with the indicated Chlamydia strains (CpoS, red; OmpA, green; Hoechst, blue; confocal images; bar=10 µm).
(B) Expression of cpoS from a plasmid vector rescues the cytotoxicity of CTL2-M007 while targeted disruption of cpoS induces host cell death in infected cells. Displayed is extracellular LDH activity (10 IFU/cell, 26 hpi).
(C) Functional diversity of Rab GTPases (Hutagalung and Novick, 2011) that co-immunoprecipitate with CpoS.
(D) Recruitment of Rab1A-EGFP to the Chlamydia inclusion depends on CpoS. HeLa cells were infected (10 IFU/cell), transfected with Rab1A-EGFP (1 hpi), and analyzed at 26 hpi (Rab1A-EGFP, white; Hoechst, green; confocal images; bar=10 µm). Arrowheads indicate inclusions.
(E) Models for the induction of cell death during infection with cpoS mutants.
(F) During co-infection of HeLa cells, CTL2-M007 inclusions (green) fuse with CTL2-R inclusions (red-left panel), but not with CTL2-M923 inclusions (red-right panel). Cells were fixed and stained (Hoechst, blue; HCS CellMask, white) at 24 hpi. Bar=20 µm.
(G) Co-infection with CTL2-R, but not CTL2-M923, protects HeLa cells from CTL2-M007-induced cell death. Infected cells (10 IFU/cell per strain) were monitored by live cell imaging from 12 to 40 hpi. At least 235 cells were analyzed per group.
Results shown represent mean±SD (n=3, one-way ANOVA + Newman-Keuls). See also Fig. S2, Tables S1–3 and movies S4–5.
We next identified CpoS-interacting proteins by expressing a FLAG-tagged CpoS in CTL2-M007, followed by non-denaturing co-immunoprecipitation with anti-FLAG antibodies and protein identification by mass spectrometry (Fig. S2E). Interacting proteins included the Inc protein CTL0476 (CT223 in C. trachomatis serovar D), consistent with predictions made by bacterial two hybrid screens (Gauliard et al., 2015), and multiple Rab GTPases, including previously reported interactions with Rab1A/B and Rab35 (Mirrashidi et al., 2015) (Fig. 2C, Table S3), suggesting that CpoS may modulate membrane trafficking. CpoS is necessary for the recruitment of a subset of these Rab proteins as Rab1A-EGFP did not associate with inclusions in cells infected with cpoS mutants (Fig. 2D).
Inclusions lacking CpoS initiate the onset of a death signal
We considered two scenarios to account for the induction of death in the absence of CpoS: (1) CpoS globally inhibits pro-death signals activated by the presence of inclusions, and (2) the presence of inclusions lacking CpoS initiates a pro-death signal (Fig. 2E). To test these scenarios we took advantage of the fusogenicity of inclusions, a process that requires the protein IncA (Hackstadt et al., 1999). Time-lapse microscopy revealed that inclusions made by CTL2-M007 expressing GFP and CTL2-R expressing mCherry readily fused (Fig. 2F, movie S4) and that the proportion of cells that died was significantly reduced compared to infection with CTL2-M007 alone (Fig. 2G). In contrast, inclusions made by the incA mutant strain CTL2-M923 (Kokes et al., 2015) (Table S1) failed to fuse with CTL2-M007 inclusions (Fig. 2F, movie S5) and to protect co-infected cells (Fig. 2G), despite the presence of CpoS on CTL2-M923 inclusions (Fig. S2F). These findings demonstrate that CpoS-defective inclusions initiate the onset of a pro-death signal that cannot be inhibited by CpoS on other inclusions. The protection conferred by CpoS is thus inclusion-autonomous.
Inhibitors of programmed cell death fail to protect cells infected with cpoS mutants
We next explored the contribution of programmed cell death pathways to the death induced by CpoS-deficient strains. The pan-caspase inhibitors Q-VD-OPH and Z-VAD-FMK failed to block CTL2-cpoS::bla-induced cell death (Fig. S3A) at concentrations that blocked apoptosis and secondary necrosis induced by staurosporine or TNF-α (Fig. S3B–C). Moreover, cell death induction occurred normally in mouse lung fibroblasts (MLFs) deficient for the pyroptotic caspases 1 and 11 (Jorgensen et al., 2011) (Fig. S3D) and could not be inhibited by two inhibitors of necroptosis, necrostatin-1 and necrosulfamide (Fig. S3E). Cell death induction was also resistant to the cathepsin B inhibitor CA-074 methyl ester and the free radical scavenger butylated hydroxyanisole (BHA) (Fig. S3F).
cpoS mutants induce an enhanced interferon response
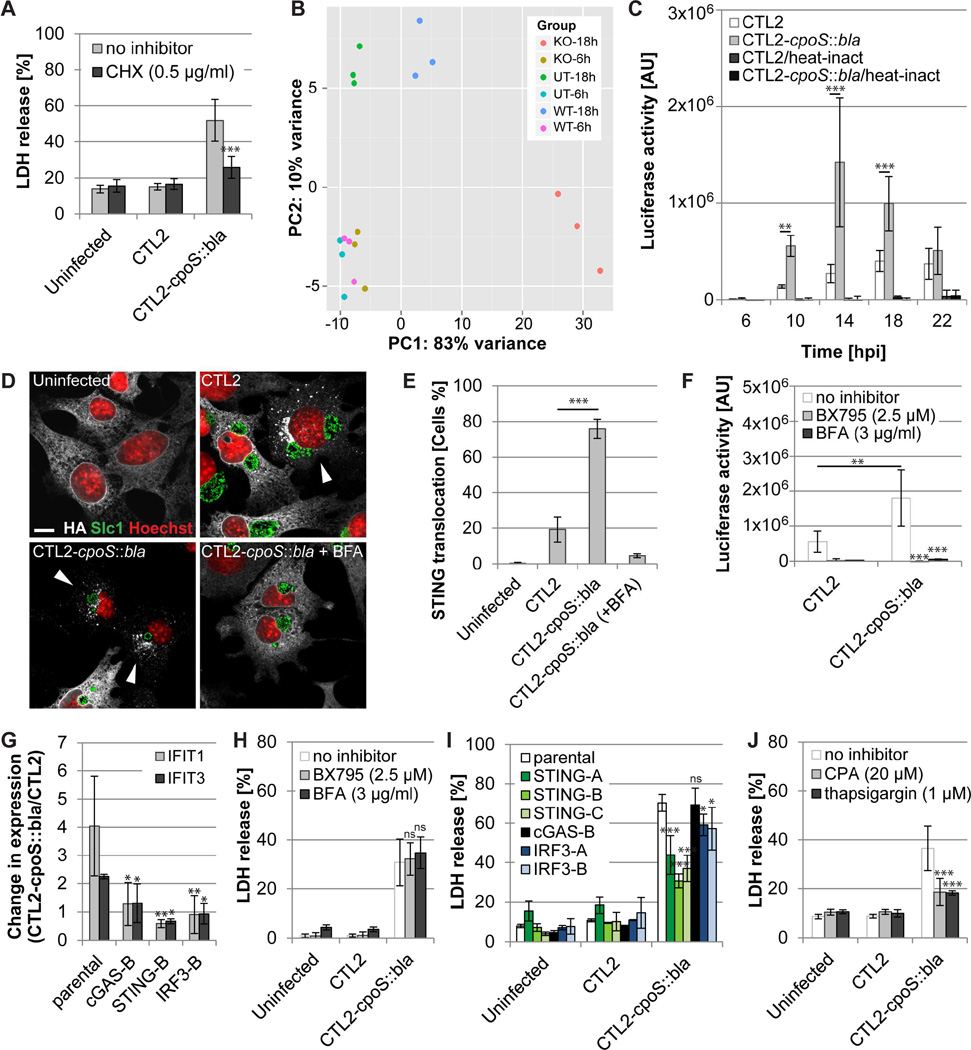
We used RNA-Seq to characterize host transcriptional responses to infection with CTL2 or CTL2-cpoS::bla, because the cell death induced by cpoS mutants was reduced by inhibitors of host protein synthesis or transcription (Fig. 3A and S4A–B). The transcriptional profile of A2EN cells infected with the mutant was strongly altered by 18 hpi, with the transcription of 400 host genes increasing by greater than two-fold as compared to CTL2-infected cells (Fig. 3B, Table S4). Gene ontology term enrichment analysis (Mi et al., 2016) and functional annotation clustering (Huang da et al., 2009) of differentially expressed genes indicated an enrichment of immunity related genes, including 27 genes encoding cytokines such as IFN-I (IFN-β), IFN-III (IFN-λ1–3), and TNF-α (Fig. S4C; Table S4). At least 58% of the upregulated genes are known IFN-I/IFN-III-stimulated genes (ISGs) (Table S4). Differential regulation was confirmed by qPCR analysis for selected targets (Fig. S4D). Consistent with the expression analysis, CTL2-cpoS::bla induced enhanced activation of an A2EN reporter cell line expressing luciferase under control of the IFN stimulated response element (ISRE); even at time points that precede the onset of cell death (i.e. 10 hpi) (Fig. 3C).
Figure 3. STING mediates type I IFN and cell death responses to cpoS mutants.
(A) Cycloheximide (CHX; added 1 h before infection) blocks CTL2-cpoS::bla-induced cell death in A2EN cells (10 IFU/cell, 24 hpi).
(B) Principal component analysis displaying differences between the transcriptional responses of uninfected A2EN cells (UT) and cells infected with CTL2 (WT) or CTL2-cpoS::bla (KO) (20 IFU/cell).
(C) Time course of luciferase activity in A2EN-ISRE reporter cells infected with the indicated strains (10 IFU/cell). Heat-inactivated (56°C, 30 min) bacteria served as control.
(D–E) Morphological (D) and quantitative (E) assessment of STING translocation in response to infection with CTL2 cpoS::bla at 18 hpi (10 IFU/cell) in Gt MLFs (expressing HA-tagged STING) by immunofluorescence microscopy (HA-tag, white; Slc1, green; Hoechst, red; confocal images; bar=10 µm). BFA (3 µg/ml, added at 6 hpi) was included as a control. Arrowheads in (D) indicate cells with evidence of STING translocation.
(F) BX795 and BFA (added at 6 hpi) inhibit type I IFN responses induced by CTL2-cpoS::bla (20 IFU/cell). Luciferase activity in A2EN-ISRE cells was measured at 14 hpi.
(G) Induction of augmented ISG (IFIT1, IFIT3) expression in HeLa cells at 14 hpi (10 IFU/cell) is dependent on STING, cGAS, and IRF3.
(H) BX795 and BFA (added at 6 hpi) fail to inhibit CTL2-cpoS::bla-induced cell death in A2EN cells (20 IFU/cell, 24 hpi).
(I) HeLa cells deficient for STING, but not IRF3 or cGAS, are partially protected from CTL2-cpoS::bla-induced cell death (10 IFU/cell, 24 hpi).
(J) SERCA inhibitors (added 1 h before infection) block CTL2-cpoS::bla-induced cell death in A2EN cells (10 IFU/cell, 24 hpi).
All experiments were conducted with purified EBs. If not stated otherwise, results shown represent mean±SD (n=3, one- or two-way ANOVA + Newman-Keuls or Sidak). See also Fig. S3 and S4 and Table S4.
STING mediates IFN and cell death responses to cpoS mutants
Induction of type I IFN during infection with Chlamydia requires the ER protein STING that is activated by cyclic dinucleotides produced by the bacteria or by the DNA senor cGAS (Barker et al., 2013; Prantner et al., 2010). Activated STING relocalizes to perinuclear vesicles to initiate TBK1 kinase-dependent phosphorylation of the transcription factor IRF3 and expression of type I interferons (Ishikawa et al., 2009; Tanaka and Chen, 2012). The frequency of cells displaying STING translocation was 3-fold higher during infection with the CpoS mutant (Fig. 3D–E) and this translocation was blocked by brefeldin A (BFA), an inhibitor of ER to Golgi protein transport (Fig. 3D–E). Activation of the ISRE-luciferase reporter was blocked by both BFA and BX795, an inhibitor of TBK1 (Fig. 3F). The augmented induction of ISG expression during infection with the CpoS mutant was abrogated in HeLa cells deficient for cGAS, STING, or IRF3 (Fig. 3G and S4E), confirming that the exacerbated IFN response induced in the absence of CpoS requires the canonical STING-dependent signaling pathway.
We expected that perturbation of interferon signaling would block the death induced by cpoS mutants. Unexpectedly, BFA and BX795 as well as cGAS and IRF3 gene deletions failed to inhibit cell death (Fig. 3H–I), suggesting that cell death and IFN responses to CpoS-deficient Chlamydia are uncoupled. However, STING-deficient HeLa cells were partially protected from cell death (Fig. 3I). A similar rescue was observed in A2EN cells in which STING was depleted by shRNA-mediated gene silencing (Fig. S4F–G) and in Goldenticket (Gt) MLFs, which have a defective allele of the STING gene (Fig. S4H). Because STING interacts with the ER calcium pump SERCA2 (Lee et al., 2013) and calcium is a regulator of cell death (Orrenius et al., 2015), we next tested the effect of compounds that alter intracellular calcium homeostasis on the cell death induced by cpoS mutants. We observed that low concentrations of thapsigargin and cyclopiazonic acid (CPA), two inhibitors of SERCA activity, both reduced the cytotoxicity of CpoS-deficient Chlamydia (Fig. 3J), providing a possible explanation for how STING mediates Chlamydia-induced cell death independent of its role in regulating interferon responses.
cpoS mutants are cleared faster from the murine genital tract
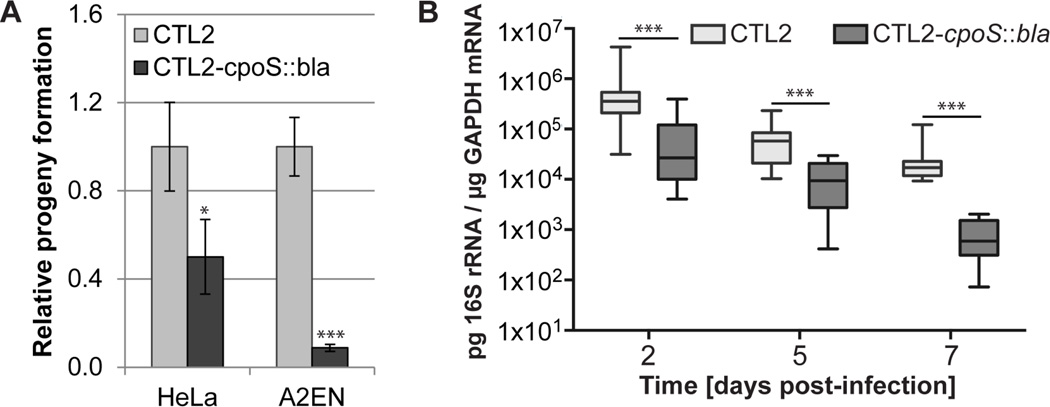
We predicted that the net effect of increased cell death and exacerbated cytokine responses in the absence of CpoS would disrupt the infectious cycle. Indeed, while primary inclusion formation was indistinguishable between CTL2 and CTL2-cpoS::bla (Fig. S4I), cpoS mutants were impaired in their ability to form infectious EBs in both HeLa and A2EN cells (Fig. 4A). When we challenged female mice transcervically, bacterial loads for the CpoS-deficient strain declined significantly faster between day 2 and 7 post-infection (Fig. 4B). Expression levels of TNF-α and GBP2 (an ISG) mirrored this trend (Fig. S4J–K). These results demonstrate that CpoS is a virulence factor that is required for optimal infection of animals.
Figure 4. cpoS mutants are defective for propagation in cell culture and in mice.
(A) CTL2 cpoS::bla produces less infectious progeny. The number of infectious bacteria was quantified at 34 hpi, normalized to the input, and displayed relative to progeny produced by CTL2 (mean±SD, n=3, student t test).
(B) CpoS-deficient bacteria are cleared faster from the mouse genital tract (box-and-whisker plot, n=13 (days 2+5), n=8 (day 7), Mann-Whitney U test).
All experiments were conducted with density gradient purified EBs. See also Fig. S4.
Discussion
Our study demonstrates that C. trachomatis actively counteracts cell death-inducing signals initiated within infected cells in response to inclusions. Inactivation of a single Inc protein was sufficient to cause the early death of the majority of cells (Fig. 2B). The observed heterogeneity in cell fate among infected cells (Fig. 1 and S1) suggests the induction of multiple pro-death signals, but could also reflect the crosstalk between signaling pathways (Vanden Berghe et al., 2015). Indeed, inhibition of any one specific death pathway often alters the mode and kinetics of cell death instead of conferring complete protection (Galluzzi et al., 2012). It is possible that our inability to block cell death by inhibiting individual pathways (Fig. S3) reflects this inherent complexity. Because the protection conferred by CpoS is restricted to the inclusion in which it resides (Fig. 2G), we consider it unlikely that this Inc is responsible for the ability of C. trachomatis to enhance resistance of infected cells to extrinsic inducers of apoptosis (Sharma and Rudel, 2009).
Chlamydia-infected cells express TNF-α and type I IFNs (Finethy and Coers, 2016), which can induce cell death or enhance the susceptibility of cells to pro-death stimuli (Malireddi and Kanneganti, 2013; Van Herreweghe et al., 2010). Yet, given that uninfected cells adjacent to cells infected with cpoS mutants were not especially prone to death (Fig. 1F) it is unlikely that the enhanced cytotoxicity was caused by enhanced induction of these cytokines. Furthermore, inhibition of TBK1 and genetic inactivation of IRF3 blocked the IFN response but not cell death (Fig. 3), indicating that cell death is not linked to either cytokines or IRF3-dependent cell death pathways described in other systems (Di Paolo et al., 2013). Nevertheless, the enhanced cytokine responses imply that host cells are sensitized to the presence of CpoS-deficient inclusions and indicate that one of the functions of CpoS is to counteract cell-autonomous defense programs by masking the presence of bacterial signals from host cells.
For some vacuolar pathogens the loss of niche integrity can trigger host cell death (Fredlund and Enninga, 2014). It is possible that CpoS preserves host cell viability by conferring stability to the inclusion. In Salmonella typhimurium and Legionella pneumophila, the effectors SifA or SdhA, respectively, counteract the action of secreted phospholipases that otherwise would compromise the integrity of the vacuole (Creasey and Isberg, 2012; Ruiz-Albert et al., 2002). Enhanced release of L. pneumophila factors from damaged vacuoles triggers type I IFN responses and cell death in macrophages (Creasey and Isberg, 2012). We consider this unlikely in our system because time-lapse microscopy of infected cells indicated that in cells undergoing necrosis the collapse of the Cpos-deficient inclusion membrane occurred either simultaneously with the rupture of the host cell membrane or only shortly before. Moreover, in cells undergoing apoptosis, inclusion integrity was preserved throughout the membrane blebbing stage (movie S3).
Because CpoS interacts with host Rab GTPases (Mirrashidi et al., 2015; Rzomp et al., 2006) (Fig. 2C–D, Table S3), and inhibition of vesicular transport blocks STING translocation (Fig. 3D–E), we predict that CpoS suppresses IFN responses by interfering with membrane trafficking events. An analogous strategy is used by Shigella flexneri to inhibit ER to ERGIC trafficking and STING signaling by targeting ARF GTPases (Dobbs et al., 2015). The specific roles of the many CpoS-interacting Rab and Inc proteins in restricting STING translocation and IFN induction remain to be determined, yet we predict that there are also non vesicular traffic-dependent activities regulated by CpoS since the contribution of STING to cell death was independent of its exit from the ER (Fig. 3H). STING localizes to ER-mitochondria contact sites (Marchi et al., 2014) and interacts with the ER calcium pump SERCA (Lee et al., 2013). Our observation that low doses of SERCA inhibitors, which deplete calcium pools in the ER (Birkett et al., 1999), partially blocked the cell death induced by cpoS mutants (Fig. 3J), suggests that STING may be a regulator of calcium-mediated cell death.
In conclusion, we provide evidence that C. trachomatis relies on CpoS to dampen the activation of cytokine and cell death pathways that control Chlamydia growth. We predict that analysis of Inc-dependent activities will reveal novel immune detection and cell protection pathways, and highlight host signaling events that can be targeted to ameliorate Chlamydia-induced pathologies.
Experimental procedures
A description of the procedures can be found in Supplemental Experimental Procedures.
Cell culture and infection
HeLa (ATCC CCL-2), Vero (ATCC CCL-81), HEK293T (ATCC CRL-3216), A2EN (Buckner et al., 2011), and THP-1 (ATCC TIB-202) cells, Gt MLFs (Barker et al., 2013), and C57BL/6 MLFs (Jorgensen et al., 2011) were maintained under standard cell culture conditions. Lentiviruses were used to generate stable silencing of STING in A2EN cells (shRNA-based) and for gene editing in HeLa cells (CRISPR/Cas9 system). C. trachomatis strain L2/434/Bu (CTL2; ATCC VR-902B), the rifampin-resistant variant CTL2-R (Nguyen and Valdivia, 2012), and derivatives described in this study were propagated in Vero cells. When indicated, infections were conducted with density gradient purified EBs (Saka et al., 2011). Cells treated with bacteria were centrifuged (1500 × g, 30 min) to enhance infection. To quantify infectious progeny formation, bacteria were harvested from infected cell monolayers at 34 hpi and used to infect fresh cell monolayers.
Generation of genetically modified Chlamydia
strains CTL2-M007 and CTL2-M923 (Kokes et al., 2015) were sequenced as described previously (Snavely et al., 2014). Chlamydia recombinants were generated by lateral gene transfer and genotyped as described previously (Nguyen and Valdivia, 2012). To enable complementation, expression of fluorescent proteins, or genetic disruption of CTL0481, Chlamydia were transformed (Wang et al., 2011) with derivatives of the E. coli-Chlamydia shuttle vector p2TK2-SW2 (Agaisse and Derre, 2013) or the TargeTron vector pDFTT3 (Johnson and Fisher, 2013).
Cell death analysis and luciferase reporter assay
For the analysis of live cells, imaging was performed with a Zeiss Axio Observer.Z1 microscope. Membrane integrity was assessed by addition of propidium iodide. Active caspase-3 was detected in live cells by incubation with NucView 488 caspase-3 substrate (Biotium) or by immunofluorescence staining with anti-active caspase-3 antibodies (BD Pharmingen). Host cell lysis and DEVD cleavage were measured using the in vitro cytotoxicity kit (Sigma-Aldrich) and the fluorimetric caspase-3 assay kit (Sigma-Aldrich), respectively. The activation of IFN-dependent genes was assessed using an A2EN-ISRE-luciferase reporter cell line generated with the Cignal Lenti ISRE Reporter (Luc) Kit (Qiagen).
Immunodetection assays
Protein extracts were generated by lysing cells in boiling 1% SDS buffer, resolved by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad). Primary rabbit antibodies used included: anti-CpoS, anti-OmpA (K. Fields, University of Kentucky), anti-FLAG (Sigma-Aldrich, F7425), anti-STING (Cell Signaling, 13647S), anti-cGAS (Cell Signaling, 15102S), and anti-IRF3 (Cell Signaling, 4302S). Primary mouse antibodies include anti-β-actin (Sigma-Aldrich, A2228). Fluorescently-labeled secondary antibodies (LI-COR Biosciences) were used and signals were recorded with an Odyssey Fc imaging system (LI-COR Biosciences). For immunofluorescence staining, primary antibodies included rabbit-anti-Slc1 (Chen et al., 2014) and anti-active-caspase-3 (BD Pharmingen, 559565), and mouse-anti-OmpA (M. Scidmore, Cornell University), rabbit-anti-CT229 (T. Hackstadt, RML/NIAID), rat-anti-HA (Sigma-Aldrich 11867423001). DNA and cells were stained with Hoechst 33258 (Life technologies) and HCS CellMask Deep Red stain (Life technologies), respectively. Images were acquired on a Zeiss Observer.Z1 epifluorescence microscope or a Zeiss 780 inverted confocal microscope.
Identification of CpoS-interacting proteins
Lysates from cells infected with CTL2-M007 expressing CpoS-FLAG or untagged CpoS were incubated with ANTI-FLAG M2 magnetic beads (Sigma-Aldrich). After immunoprecipitation bound proteins were eluted and separated by SDS-PAGE. LC-MS/MS was used to identify proteins enriched in samples from cells infected with CpoS-FLAG expressing bacteria. For validation, HeLa cells were transfected with a Rab1A-EGFP expression plasmid (Rzomp et al., 2003).
Murine infection model
Female C57BL/6 mice (Jackson Laboratory) were treated with 2.5 mg medroxyprogesterone subcutaneously. Seven days later, mice were infected transcervically with 1 × 107 EBs per mouse (Coers et al., 2011). Mice were sacrificed and upper genital tract samples were immediately homogenized. RNA was isolated by TRIzol (Thermo Fisher Scientific) extraction.
RNA-Seq and RT-qPCR
Total RNA was isolated using the RNeasy Mini Plus kit (Qiagen). Sequencing libraries, prepared using the Kapa Stranded mRNA-Seq kit (Kapa Biosystems) were pooled and sequenced together on a total of 3 lanes of Illumina HiSeq 2000/2500 to obtain 50 bp single-end reads that were processed, mapped, and counted. Normalization and differential expression analysis were carried out using the DESeq2 R package (Love et al., 2014). A cutoff FDR (False Discovery Rate) of less than 0.001 and a log fold change of more than 1 were used to select significantly differentially expressed genes. RT-qPCR was conducted using the Power SYBR Green RNA-to-CT™ 1-Step Kit (Thermo Fisher Scientific). PCR targets and primers are listed in Supplemental Experimental Procedures.
Statistics
If not stated otherwise, statistical analysis was performed using the software GraphPad Prism 6.01 (*, p<0.05; **, p<0.01; p<0.001; ns, not significant).
Supplementary Material
Highlights.
A genetic screen identifies C. trachomatis effector CpoS as a regulator of cell death
Loss of CpoS on the inclusion causes enhanced death in infected cells
DNA sensor STING modulates IFN responses and death in cells infected with cpoS mutants
cpoS mutants are defective for propagation in cell culture and in mice
Acknowledgments
We thank the Duke University Core facilities for their technical support and T. Hackstadt (RML/NIAID), M. Scidmore (Cornell University), K. Fields (University of Kentucky), I. Dérre (University of Virginia), H. A. Saka (National University of Cordoba), D. J. Fisher (Southern Illinois University), and K. Spaeth for sharing cell lines, plasmids and reagents. This work was supported by the European Union's Seventh Framework Program (grant n°PIOF-GA-2013-626116), the National Institute of Health (NIH) (R01AI100759, R01AI103197), a Burroughs Wellcome Fund PATH Award, and a National Science Foundation (NSF) predoctoral award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, B.S.S. and R.H.V.; Investigation, B.S.S., R.J.B., R.F., and R.M.B.; Writing – Original Draft, B.S.S. and R.H.V.; Resources, V.K.C.; Supervision, G.K., J.C., and R.H.V.
References
- Agaisse H, Derre I. A C trachomatis cloning vector and the generation of C trachomatis strains expressing fluorescent proteins under the control of a C trachomatis promoter. PLoS One. 2013;8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio. 2013;4:e00018–e00013. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkett SD, Jeremy JY, Watts SM, Shukla N, Angelini GD, McArdle CA. Inhibition of intracellular Ca2+ mobilisation by low antiproliferative concentrations of thapsigargin in human vascular smooth-muscle cells. J Cardiovasc Pharmacol. 1999;33:204–211. doi: 10.1097/00005344-199902000-00005. [DOI] [PubMed] [Google Scholar]
- Buckner LR, Schust DJ, Ding J, Nagamatsu T, Beatty W, Chang TL, Greene SJ, Lewis ME, Ruiz B, Holman SL, et al. Innate immune mediator profiles and their regulation in a novel polarized immortalized epithelial cell model derived from human endocervix. J Reprod Immunol. 2011;92:8–20. doi: 10.1016/j.jri.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 2014;10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Gondek DC, Olive AJ, Rohlfing A, Taylor GA, Starnbach MN. Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections. PLoS Pathog. 2011;7:e1001346. doi: 10.1371/journal.ppat.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci U S A. 2012;109:3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo NC, Doronin K, Baldwin LK, Papayannopoulou T, Shayakhmetov DM. The transcription factor IRF3 triggers "defensive suicide" necrosis in response to viral and bacterial pathogens. Cell Rep. 2013;3:1840–1846. doi: 10.1016/j.celrep.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Coers J. Sensing the enemy, containing the threat: cell-autonomous immunity to Chlamydia trachomatis . FEMS Microbiol Rev. 2016 doi: 10.1093/femsre/fuw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund J, Enninga J. Cytoplasmic access by intracellular bacterial pathogens. Trends Microbiol. 2014;22:128–137. doi: 10.1016/j.tim.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard E, Ouellette SP, Rueden KJ, Ladant D. Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Front Cell Infect Microbiol. 2015;5:13. doi: 10.3389/fcimb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Bednar MM, Amin V, Davis BK, Ting JP, McCafferty DG, Valdivia RH. The Chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe. 2011;10:21–32. doi: 10.1016/j.chom.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K, Saleh M. Cell death in the host response to infection. Cell Death Differ. 2008;15:1339–1349. doi: 10.1038/cdd.2008.91. [DOI] [PubMed] [Google Scholar]
- Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, Dotiwala F, Sen J, Doench JG, Orzalli MH, et al. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14:179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malireddi RK, Kanneganti TD. Role of type I interferons in inflammasome activation, cell death, and disease during microbial infection. Front Cell Infect Microbiol. 2013;3:77. doi: 10.3389/fcimb.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim Biophys Acta. 2014;1837:461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, et al. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas I. Female genital Chlamydia trachomatis infection: where are we heading? Arch Gynecol Obstet. 2012;285:1271–1285. doi: 10.1007/s00404-012-2240-7. [DOI] [PubMed] [Google Scholar]
- Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A. 2012;109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S, Gogvadze V, Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem Biophys Res Commun. 2015;460:72–81. doi: 10.1016/j.bbrc.2015.01.137. [DOI] [PubMed] [Google Scholar]
- Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. Proteins in the chlamydial inclusion membrane. Microbes Infect. 2002;4:333–340. doi: 10.1016/s1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Albert J, Yu XJ, Beuzon CR, Blakey AN, Galyov EE, Holden DW. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun. 2006;74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol. 2011;82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Rudel T. Apoptosis resistance in Chlamydia-infected cells: a fate worse than death? FEMS Immunol Med Microbiol. 2009;55:154–161. doi: 10.1111/j.1574-695X.2008.00515.x. [DOI] [PubMed] [Google Scholar]
- Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis. 2014;71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herreweghe F, Festjens N, Declercq W, Vandenabeele P. Tumor necrosis factor-mediated cell death: to break or to burst, that's the question. Cell Mol Life Sci. 2010;67:1567–1579. doi: 10.1007/s00018-010-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Kaiser WJ, Bertrand MJ, Vandenabeele P. Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol Cell Oncol. 2015;2:e975093. doi: 10.4161/23723556.2014.975093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ME. The chlamydial developmental cycle. In: Barron AL, editor. Micobiology of Chlamydia. Boca Raton FL: CRC Press; 1988. pp. 71–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.