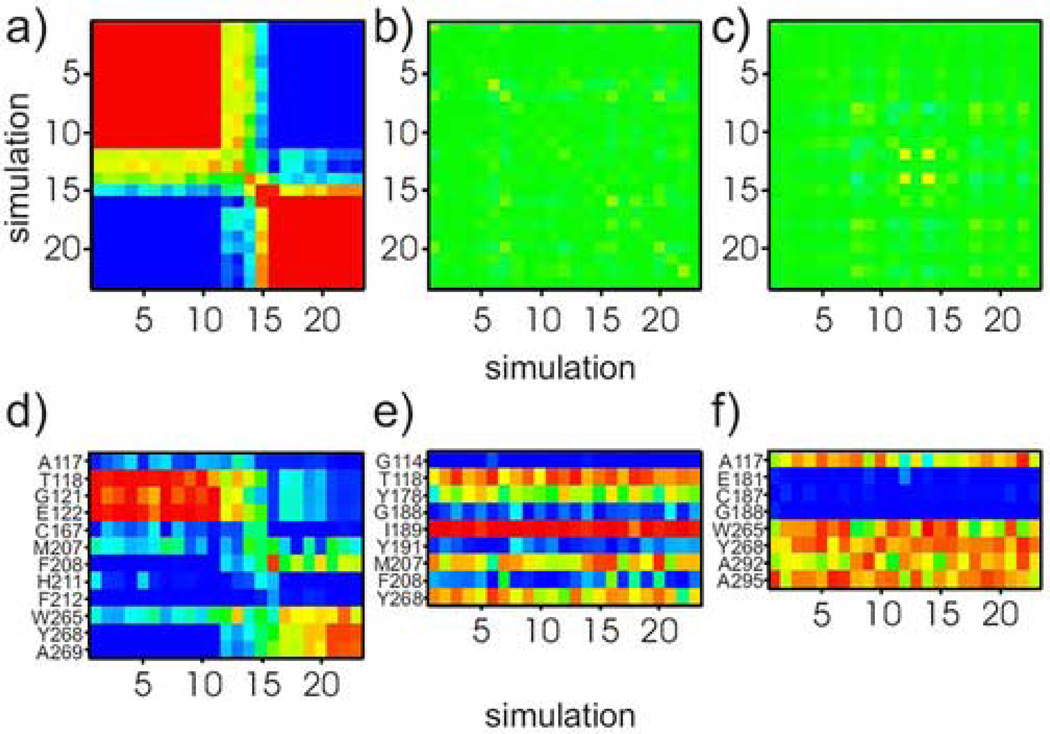
Figure 5.
Covariance analysis of individual 100-ns simulations shows variability of protein environment of retinylidene C5-, C9-, and C13-methyl groups. Elements of the covariance matrix are depicted by colors and range from −1 (completely anti-correlated; blue) to 1 (completely correlated; red). Panels (a)–(c) correspond to C5-, C9- and C13-methyl positions, respectively. Note that the matrix in panel (a) has three different blocks: (i) simulations 1– 11, (ii) simulations 12– 15, and (iii) simulations 16–23. Each of the blocks has a distinct C5-methyl environment. By contrast, in (b) and (c) the color map for the C9-, and C13-methyls has a consistent green color, indicating all 23 simulations yield similar methyl group environments. Panels (d)–(f) show contact matrices for the C5-, C9-, and C13-methyl groups, respectively. Columns list the simulation number and rows correspond to residues within a 4 Å-radius sphere centered on the methyl carbon. The color scale ranges from blue (low fraction value; indicating residue is infrequent within 4 Å-radius sphere) to red (high fraction showing sampling of different environments. For the C9- and C13-methyls in (b) and (c), bands of similar colors across all 23 simulations evince similar environments.