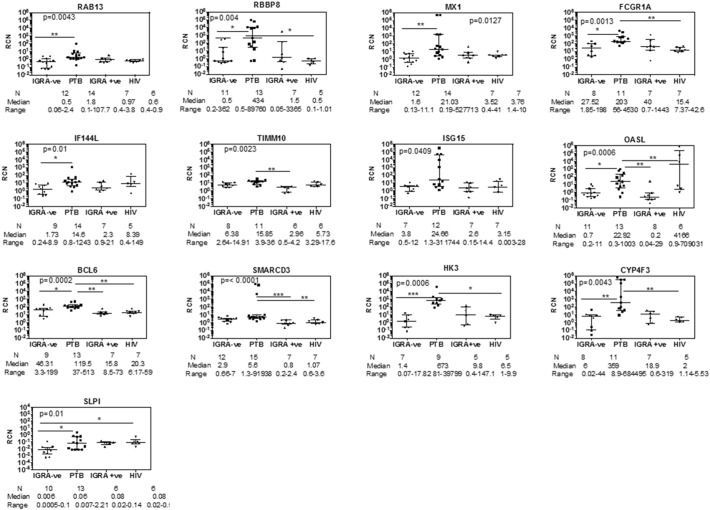
Fig. 6.
qRT-PCR verification of the 16 genes shortlisted by the biomarker identification pipeline. Comparative analysis of IGRA ve/healthy controls, active pulmonary TB, latent TB and HIV infected subjects: All genes listed were verified by qRT-PCR by the SYBR method, with the exception of SLPI, which was verified by TaqMan. GAPDH was used as internal control for calculating relative copy number (RCN) of each gene. Statistical significance for between group RCN differences was first calculated using Mann–Whitney U test between IGRA − ve and active TB subjects. All genes tested were significantly different between these two groups using this test. p-Value shown in each graph was determined by one way ANOVA to correct for multiple comparisons; Dunn's multiple comparisons test was then used to identify significant differences between specific groups as shown by a line. *p = 0.01; **p < 0.01, ***p < 0.0001.