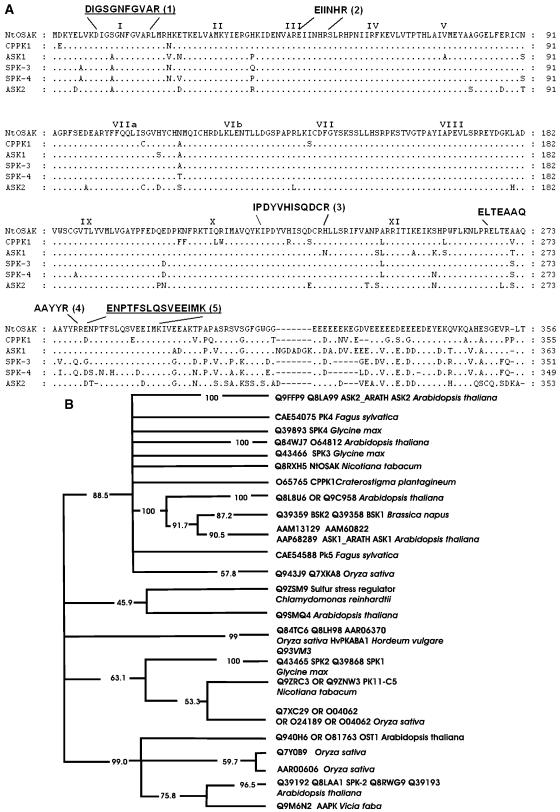
Figure 1.
NtOSAK belongs to the SnRK2 subfamily. A, Comparison of the predicted amino acid sequence of NtOSAK with sequences of other protein kinases. The sequences of five protein kinases exhibiting sequence similarity are aligned with that of NtOSAK. References for the sequences of the listed kinases are as follows: CPPK1-protein kinase 1 from Craterostigma plant (P. Heino, M. Nylander, T. Palva, and D. Bartels, unpublished data; Swiss-Prot accession no. O65765); soybean protein kinases SPK-3 (Swiss-Prot accession no. Q43466) and SPK-4 (Swiss-Prot accession no. Q39893; Yoon et al., 1997); and Arabidopsis protein kinases ASK1 (Swiss-Prot accession no. P43291) and ASK2 (Swiss-Prot accession no. P43292; Park et al., 1993). The conserved Ser/Thr protein kinase subdomains (Hanks et al., 1988) are indicated by roman numerals. Gaps are marked with dashes and were introduced to maximize alignment. The peptide sequences obtained by microsequencing are shown above the NtOSAK sequence. The peptide sequences used for designing PCR primers are underlined. B, Phylogenetic tree of plant SNF1-related protein kinases that are closely related to NtOSAK. The tree is a consensus of 100 maximum parsimony “bootstrap” trees. Percentage values on each branch represent the corresponding bootstrap probabilities. The multiple alignment was created using the CLUSTALW computer program (Thompson et al., 1994). Columns with gaps and places where alignment seemed doubtful were excluded. The multiple alignment after edition contains 282 columns. The consensus maximum parsimony tree was calculated using the PHYLIP package (Felsenstein, 1989).