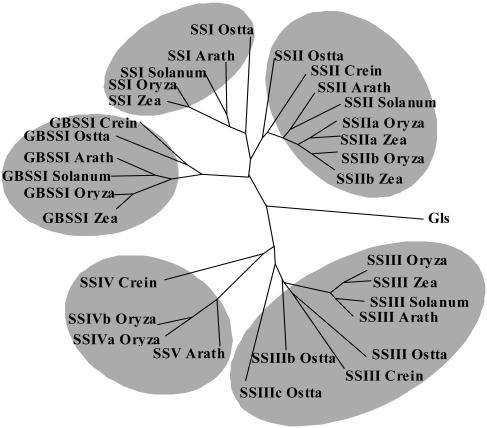
Figure 5.
Dendogram of starch synthases in plants and algae and glycogen synthases from Synechocystis. Sequences were aligned with ClustalW (Thompson et al., 1994). The sequence alignment was manually improved using BioEdit (Hall, 1999) and ForCon (Van de Peer et al., 1998). TreeCon (Van de Peer and De Wachter, 1997a, 1997b) was used for constructing the neighbor-joining (Saitou and Nei, 1987) tree based on Poisson-corrected distances, only taking into account unambiguously aligned positions (600 amino acids). Bootstrap analysis with 500 replicates was performed to test the significance of the nodes. The amino acid sequences used were as follows: C. reinhardtii (Crein) GBSSI (AF26420), SSII (AAC17970), SSIII, SSIV (TO7926); maize (Zea) GBSSI (M24258), SSI (AF036891), SSIIa (Harn et al., 1998), SSIIb (Harn et al., 1998); rice (Oryza) GBSSI (X62134), SSI (D16202), SSIIa (AF419099), SSIIb (Harn et al., 1998), SSIII (Gao et al., 1998), SSIVa (AY1OO470), SSIVb (AY100471); Solanum tuberosum (Solanum) GBSSI (X58453), SSI (Y10416), SSII (X87988), SSIII (X94400); Arabidopsis (Arath) GBSSI (AC006424), SSI (AF121673), SSII (AC008261), SSIII (AC007296), SSV (A021713), and Synechocystis glycogen-synthase Gls (NP441947). Accession numbers for Ostreococcus sequences are given as supplemental material.