Abstract
P-HYDROXYPHENYLPYRUVATE DIOXYGENASE (HPPD) is the first committed enzyme involved in the biosynthesis of vitamin E, and is characterized by catalyzing the conversion of p-hydroxyphenyl pyruvate (HPP) to homogentisic acid (HGA). Here, an HPPD gene was cloned from Medicago sativa L. and designated MsHPPD, which was expressed at high levels in alfalfa leaves. PEG 6000 (polyethylene glycol), NaCl, abscisic acid and salicylic acid were shown to significantly induce MsHPPD expression, especially in the cotyledons and root tissues. Overexpression of MsHPPD was found to significantly increase the level of β-tocotrienol and the total vitamin E content in Arabidopsis seeds. Furthermore, these transgenic Arabidopsis seeds exhibited an accelerated germination time, compared with wild-type seeds under normal conditions, as well as under NaCl and ABA treatments. Meanwhile, the expression level of several genes associated with ABA biosynthesis (NCED3, NCED5 and NCED9) and the ABA signaling pathway (RAB18, ABI3 and ABI5) were significantly down-regulated in MsHPPD-overexpressing transgenic lines, as well as the total free ABA content. Taken together, these results demonstrate that MsHPPD functions not only in the vitamin E biosynthetic pathway, but also plays a critical role in seed germination via affecting ABA biosynthesis and signaling.
Vitamin E is an essential nutrient for animals and humans, the physiological significance of this substance has been studied widely. Specifically, vitamin E has been shown to scavenge singlet oxygen1, reduce lipid oxidation by-products and inhibit lipid peroxidation2, thereby helping plants to defend against various stresses and extending the shelf life of meat via keeping it fresh3. Furthermore, vitamin E deficiency has been shown to result in infertility and fetal death in animals4,5. In contrast, sufficient uptake of vitamin E in human and animal diets leads to numerous benefits, such as a decreased risk of select cancers and atherosclerosis, a bolstering of the immune system, and a reduction in instances of various vision maladies6.
Vitamin E is not a single compound, but rather the collective name for a group of eight lipid-soluble antioxidants consisting of a polar chromanol head group and a hydrophobic prenyl tail7, which are derived from the methylerythritol-4-phosphate (MEP) and shikimate pathways. Four of these compounds are termed tocopherols, and the other four are termed tocotrienols, and depending on the saturation level of the hydrophobic tail and also the number and position of the methyl groups on the chromanol ring, members of the vitamin E group are classified into α-, β-, γ- and δ- forms8. The plastidial aromatic amino acid metabolism pathway is utilized for the synthesis of the tocopherol and tocotrienol head group (homogentisic acid, HGA) and the deoxyxylulose-5-phosphate pathway is used for the synthesis of the hydrophobic tail (either phytyl-PP for the tocopherols or GGDP for the tocotrienols). Vitamin E is only synthesized in higher plants and other oxygen-evolving phototrophs, including some cyanobacteria and all species of green algae9. In plants, the production of HGA is the first step in tocopherol synthesis, and HGA is synthesized from p-hydroxyphenyl pyruvate (HPP) in a reaction catalyzed by HPPD. Ergo, HPPD is essential for plant viability, and mutant plants with null alleles of HPPD exhibit a lethal photobleaching phenotype10. It has also been shown previously that overexpression of barley HPPD results in a two-fold increase to the vitamin E content of transgenic tobacco seeds11. Furthermore, overexpression of AtHPPD increased the total vitamin E level by seven-fold in Synechocystis12, and significantly increased vitamin E accumulation in transgenic potato tubers13. Collectively, these observations indicate that modifying HPPD gene expression is a valid strategy to utilize during attempts to modulate the total vitamin E content of plant tissues.
The growth inhibitor abscisic acid (ABA) is widely recognized as an important phytohormone involved in plant stress response and seed germination14. It accumulates most notably in dry seeds and declines rapidly subsequent to seed germination15. ABA is formed via the cleavage of C40 carotenoids, which originate from the MEP pathway16,17 and share a common condensed GGDP intermediate with vitamin E. Isopentenyl pyrophosphate (IPP), which is synthesized from the MEP and/or Mevalonate (MVA) pathways, is subjected to a cascade of reactions, before ultimately being condensed to form geranylgeranyl diphosphate (GGDP), which has been shown to be a key intermediate in the synthesis of carotenoids and tocochromanols18. With regards to ABA, 9-cis-neoxanthin, which is formed during the course of the carotenoid biosynthetic pathway, is cleaved by 9-cis-epoxycarotenoid dioxygenase (NCED) genes to form xanthoxin, which is ultimately modified to ABA16. Nine NCED genes have been identified in Arabidopsis, amongst which NCED 2, 3, 5, 6, and 9 are thought to play principal roles in determining the ABA content19. There are also a number of genes involved in the ABA signaling pathway, and ABSISIC ACID INSENSITIVE 3 (ABI3) and ABSISIC ACID INSENSITIVE 5 (ABI5) have been shown to play important roles pertaining to seed germination20,21.
Alfalfa, as an important perennial leguminous forage crop, has multiple agro-ecological advantages over other crop plants, including protecting soil from erosion, as well as fixing and providing nitrogen for neighboring plants22,23. Additionally, alfalfa is considered an important feedstock that provides vitamins, proteins, and minerals to animals. However, alfalfa is a perennial autotetraploid, and its cross-pollinated genetic background has classically restricted the discovery and application of novel gene resources in alfalfa24. Vitamin E biosynthesis has been widely studied in other plants; however, little is known about the genes involved in vitamin E biosynthesis in forage crops. As such, discovering and characterizing related genes will enrich our knowledge on the biosynthetic mechanisms of this essential nutrient in forage crops, including alfalfa.
In this study, we identified an HPPD gene in alfalfa, determined that that it is phylogentically closest to MtHPPD from Medicago truncatula. The expression of MsHPPD was induced by different stress conditions in alfalfa, and overexpression of MsHPPD increased the vitamin E content in Arabidopsis seeds. The germination of these transgenic Arabidopsis seeds was accelerated, as compared to wild type seeds, under normal growth conditions. Moreover, seeds from transgenic Arabidopsis were more resistant to salt stress and less sensitive to ABA treatment, manifesting in the transgenic seeds having an elevated germination rate. Ultimately, in addition to its role in vitamin E accumulation, we showed that MsHPPD plays a positive role in seed germination via regulating ABA biosynthesis and the subsequent ABA signaling pathway.
Results
Cloning and sequence analysis of the MsHPPD gene from Medicago sativa
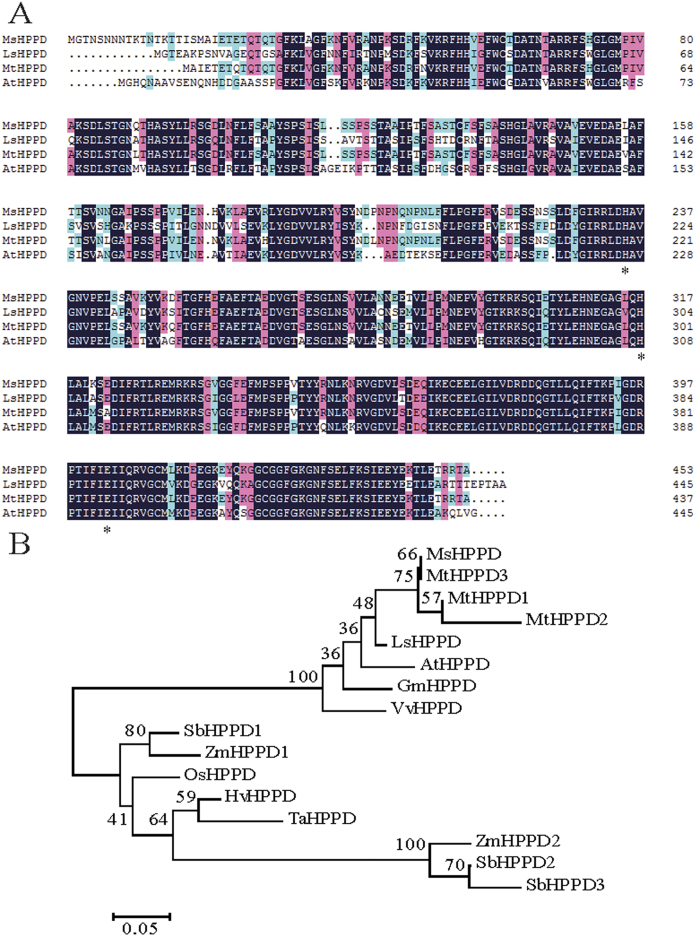
Using the known sequence of Medicago truncatula HPPD gene, a conserved 648-bp fragment was cloned from alfalfa. Rapid amplification of cDNA ends (RACE) was performed based on this conserved sequence, and a 2064-bp full-length HPPD gene was obtained by combining the1188-bp and 1001-bp fragments isolated by 3′ RACE and 5′ RACE, respectively. This full-length sequence contains a 1305-bp open reading frame, which encodes a protein of 434 amino acids. Multiple protein sequence alignment revealed that the protein sequence was most closely related to known HPPD protein sequences from other organisms, which belong to the Glo_EDI_BRP_ like superfamily, and contain an HPPD-N-like and an HPPD-C-like domain. Additionally, three iron (Fe2+) binding sites, which are essential for the activity of HPPD, are also conserved among all the HPPD sequences (Fig. 1A), including in the alfalfa sequence. Thus, the identified gene from alfalfa was designated MsHPPD (accession number KY081399). To investigate the evolutionary relationships between MsHPPD and HPPD homologs from other species, phylogenetic analyses were performed with MEGA6.0 using the Neighbor-joining method25. This showed that HPPDs from monocotyledonous and dicotyledonous species grouped into two separate clades. As expected, MsHPPD clusters together with the eudicotyledonous sequences, including MtHPPD from Medicago truncatula, LsHPPD from Lactuca sativa, and AtHPPD from Arabidopsis thaliana (Fig. 1B).
Figure 1. Bioinformatic analysis of the MsHPPD sequence.
(A) Multiple sequence alignment of plant HPPDs. MsHPPD: Medicago sativa; LsHPPD: Lactuca sativa (ACN78586.1); MtHPPD: Medicago truncatula (XP_003617391.1); AtHPPD: Arabidopsis thaliana (CBI85437.1). Asterisk: Fe binding sites. Colors highlighted homology levels. Black represents identity =100%, red represents identity ≥75%, green represents identity ≥50%. (B) Phylogenetic tree of MsHPPD and HPPDs from other plant species. MEGA 6.0 was used to construct the tree using the neighbor-joining method, bootstrap =1000. Protein sequences of HPPD were downloaded from NCBI as follows: MtHPPD1: Medicago truncatula (XP_003617384.1); MtHPPD2: Medicago truncatula (XP_003617382.2); MtHPPD3: Medicago truncatula (XP_003617391.1); LsHPPD: Lactuca sativa (ACN78586.1); AtHPPD: Arabidopsis thaliana (CBI85437.1); VvHPPD: Vitis vinifera (CAN71143.1); GmHPPD: Glycine max (ABQ96868.1); OsHPPD: Oryza sativa (EAZ21880.1); ZmHPPD1: Zea mays: (NP_001105782.1); ZmHPPD2: Zea mays: (XP_008653702.1); SbHPPD1: Sorghum bicolor (XP_002453359.1); SbHPPD2: Sorghum bicolor (XP_002461829.1); SbHPPD3: Sorghum bicolor (XP_002461838.1); TaHPPD: Triticum aestivum (CAJ29893.1); HvHPPD: Hordeum vulgare (CBI85441.1).
MsHPPD expression in different tissues
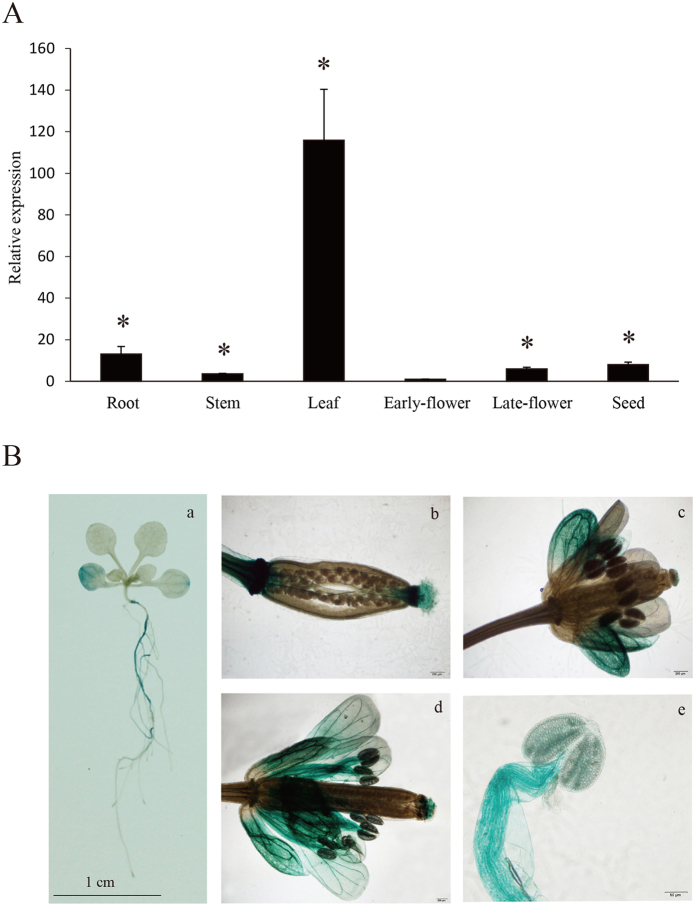
Quantitative reverse transcription-PCR (qRT-PCR) was performed to determine the expression pattern of MsHPPD in different organs of M. sativa. The results showed MsHPPD transcripts were detectable in all tested organs, with the highest expression in rosette leaves and the lowest expression in early flowers (Fig. 2A). This implies that MsHPPD may play a more active role in leaves.
Figure 2. Expression pattern analysis of MsHPPD gene.
(A) Expression levels of MsHPPD in different organs of alfalfa. RNA was extracted from tissues collected from two-year-old alfalfa. Data presented are mean ± SD, each with three biological replicates and three technical replicates. Expression levels are relative to early flower; Statistical analyses were carried out via a two-tailed Student’s t-test, asterisks show the significance of P < 0.05. (B) Histochemical GUS staining of pHPPD::GUS lines. (a) Ten-day-old Arabidopsis seedling, bar: 1 cm; (b) Silique of transgenic Arabidopsis, bar: 200 μm; (c) Early flower, bar: 200 μm; (d) Late flower, bar: 200 μm; (e) Pollen, bar: 50 μm.
To further confirm the transcriptional expression pattern of MsHPPD, a pHPPD::GUS construct was transformed into Arabidopsis. GUS expression was detected in cotyledons, primary roots, sepals, petals, stigma, filament tubes, pollens, and the ends of seeds in transgenic lines. However, no GUS activity was observed in the root tip or hypocotyls (Fig. 2B).
MsHPPD expression under various conditions
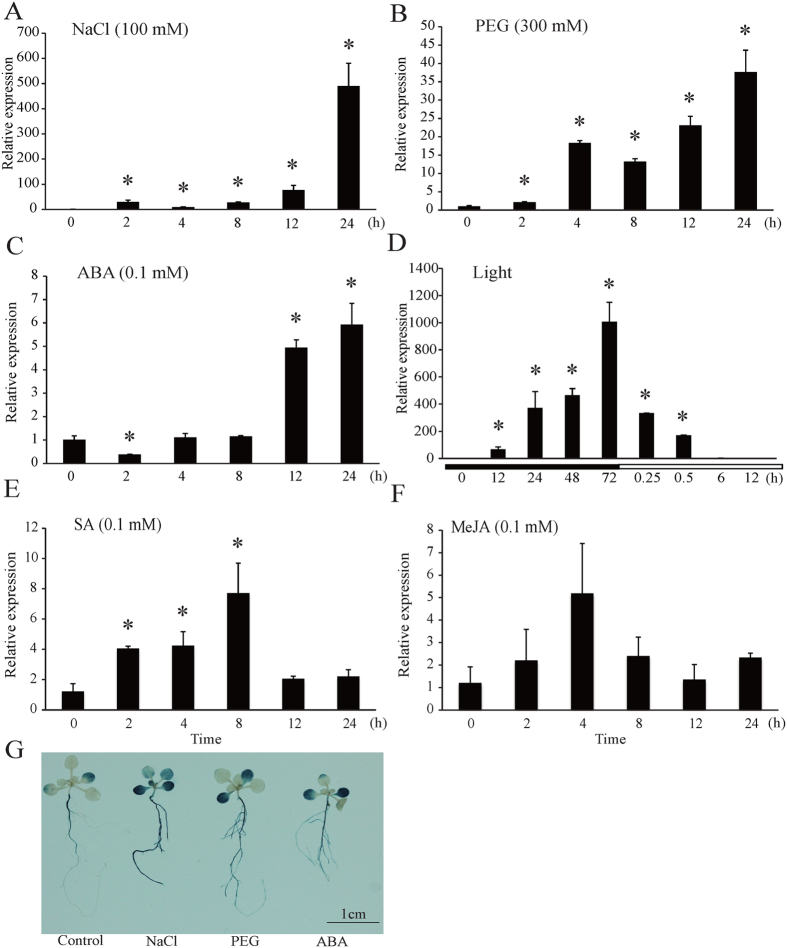
The promoter sequence of MsHPPD was analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to better investigate the expressional regulation of MsHPPD26. The results revealed cis-elements, which have been shown to respond to defense and stress signals including salicylic acid, gibberellin, and light signals are presented in the promoter region of MsHPPD (Fig. S1). To explore the possible roles of these cis-elements with regard to regulating the expression of MsHPPD, expressional analysis of MsHPPD was executed under various stress treatments. These results showed that MsHPPD transcripts were gradually up-regulated in seedlings treated with increasing treatment period when seedlings were exposed to NaCl, PEG, and ABA (Fig. 3A–C). Most notably, the expression of MsHPPD increased 500-fold after a 24 h treatment with NaCl, relative to the levels seen in the mock treatment. Likewise, treatments utilizing extended periods of darkness revealed that the expression of MsHPPD rose gradually as the darkness treatment time increased; however, upon exposure to light, the expression levels of MsHPPD dropped sharply, and this decaying trend continued with prolonged exposure to light (Fig. 3D). The expression levels of MsHPPD increased moderately, albeit significantly, subsequent to salicylic acid treatment, until the 8 h time point, at which point MsHPPD expression returned to the control level (Fig. 3E). No significant differences were observed after methyl jasmonic acid treatment (Fig. 3F).
Figure 3. Expression profiles of MsHPPD under various treatments.
(A) Expression levels of MsHPPD under NaCl treatment. (B) Expression levels of MsHPPD under PEG treatment. (C) Expression levels of MsHPPD under ABA treatment. (D) Expression levels of MsHPPD under dark to light transduction treatment. (E) Expression levels of MsHPPD under SA treatment. (F) Expression levels of MsHPPD under MeJA treatment. Two-week-old alfalfa seedlings were treated with NaCl (100 mM), PEG (300 mM), ABA (0.1 mM), dark/light, SA (0.1 mM) and MeJA (0.1 mM). Samples were collected at each time point, with three biological replicates. Data are presented as mean ± SD, each with three biological replicates and three technical replicates. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05. (G) Histochemical GUS staining of pHPPD::GUS under various conditions. Two-week-old Arabidopsis seedlings were treated with NaCl (100 mM), PEG (300 mM) and ABA (0.1 mM) for 24 h and then stained with GUS. Bar: 1 cm.
pHPPD::GUS was used to approximate the expression levels of MsHPPD in response to NaCl, PEG, and ABA treatments. Seedlings treated with NaCl, PEG, or ABA showed increase GUS activity across the plants, especially in the roots and cotyledons. This observation is consistent with the qRT-PCR results (Fig. 3G).
Expression of MsHPPD and endogenous vitamin E pathway genes in transgenic Arabidopsis
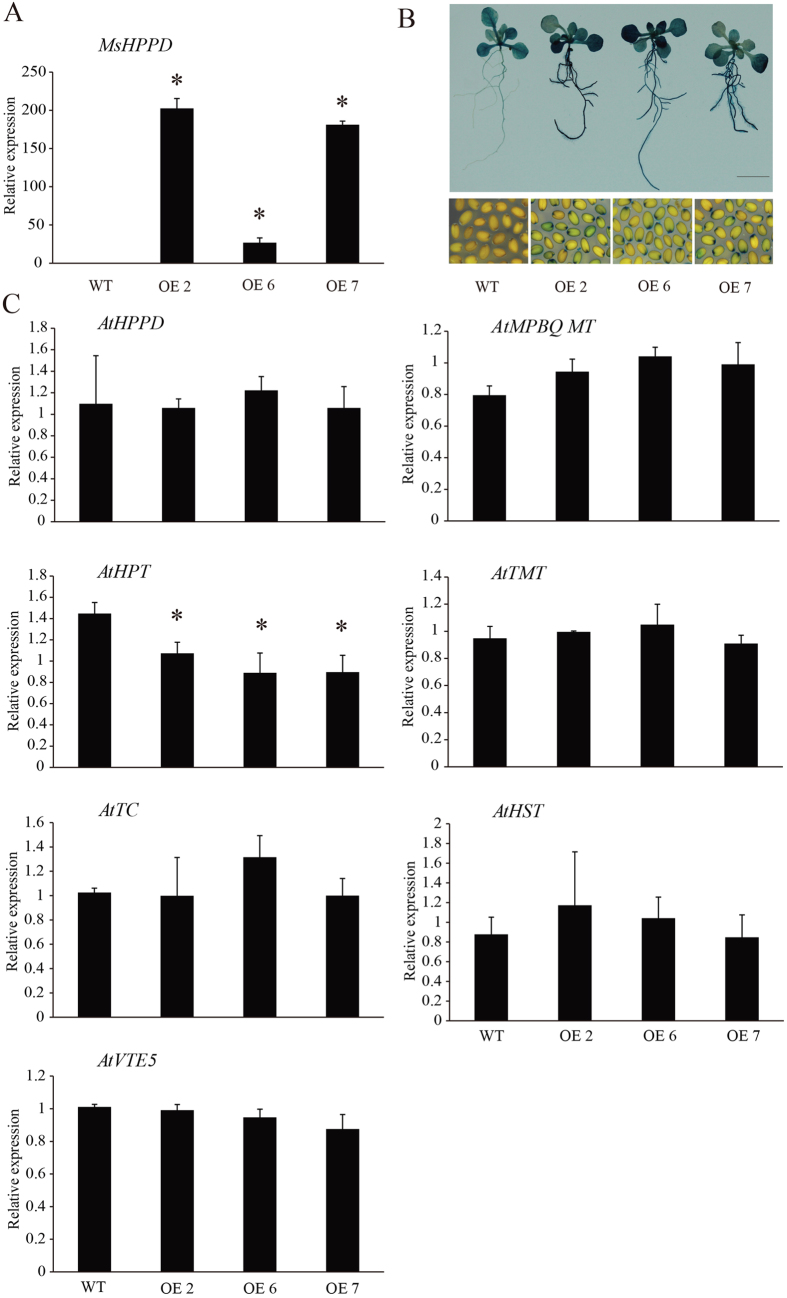
qRT-PCR was used to examine the expression levels of MsHPPD in MsHPPD-overexpressing lines and wild-type Arabidopsis. This data showed that MsHPPD was more-highly expressed in transgenic Arabidopsis lines, to varying degrees, as compared to wild-type expression levels (Fig. 4A). Likewise, all of the pHPPD::GUS transgenic Arabidopsis lines exhibited higher levels of GUS activity compared with the corresponding wild-type plants (Fig. 4B), which was consistent with the transcriptional levels of MsHPPD. To test whether the overexpression of exogenous MsHPPD alters the endogenous vitamin E biosynthetic pathway, the expression levels of critical endogenous vitamin E pathway genes were quantified. There are several genes that have been shown to regulate vitamin E biosynthesis. HPPD and HOMOGENTISATE PHYTYLRANSFERASE (HPT) have been found to play roles pertaining to determining the total vitamin E content, while 2-METHYL-6-PHYTYLBENZOQUINONEMETHYLTRANSFERASE (MPBQMT) and γ-TOCOPHEROL METHYL TRANSFERASE (γ-TMT) play roles in determining the vitamin E composition18. A significant decrease of AtHPT, which plays a role in determining vitamin E content and is also the first gene downstream of HPPD in vitamin E biosynthesis, was observed in the generated MsHPPD-overexpressing lines, but the expression levels of AtMPBQMT, AtTMT, AtTC, AtHPPD and AtVTE5 were not in the wild-type. Since HPPD is also involved in plastoquinone-9 and plastochromanol-8 biosynthesis, the expression level of AtHST, which is the first gene downstream of HPPD in plastoquinone-9 and plastochromanol-8 biosynthesis, was examined, and no significant difference was observed. This suggests that the plastoquinone-9 and plastochromanol-8 biosynthetic pathway is not altered (Fig. 4C).
Figure 4.
(A) MsHPPD expression in three MsHPPD-overexpressing transgenic lines and wild-type, as measured by qRT-PCR. Total RNA was extracted from two-week-old MsHPPD-overexpressing transgenic Arabidopsis (OE2, OE6, and OE7) and control seedlings, before being subjected to qRT-PCR analysis. The expression levels of MsHPPD were normalized to At4g26410. Data are presented as mean ± SD, each with three biological replicates and three technical replicates. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05. (B) Histochemical GUS staining of MsHPPD-overexpressing lines (OE2, OE6, and OE7) and wild-type seedlings with pBI121 empty vector in the Col-0 background. Two-week-old transgenic and control Arabidopsis were used for GUS staining. Bar: 1 cm. (C) Expression analysis of endogenous genes involved in vitamin E biosynthesis in MsHPPD- overexpressing Arabidopsis (OE2, OE6, and OE7) and wild-type control. Two-week-old Arabidopsis seedlings were used for qRT-PCR. Data are shown as the mean ± SD with three biological replicates and three technical replicates. The expression levels of the genes detected in the experiment were normalized to At4g26410. Statistical analyses were carried out via a two-tailed Student’s t tests with a significance of P < 0.05.
Analysis of vitamin E levels in transgenic Arabidopsis seeds and leaves
To test whether MsHPPD is a functional gene involved in the production of tocotrienols and tocopherols, all vitamin E forms were quantified in the seeds and leaves of MsHPPD-overexpressing lines and wild-type Arabidopsis. β-tocotrienol and δ-tocopherol content were found to have increased by 12–36% and 10–57%, respectively, in transgenic Arabidopsis seeds compared to wild-type seeds (Fig. 5). However, δ-tocotrienol was undetectable in all lines and the content of the other forms of vitamin E was not altered (Fig. S2A). Total vitamin E content was significantly higher in the seeds of MsHPPD-overexpressing lines OE2 and OE7 than the corresponding content in the seeds of the control line (Fig. 5). This corresponds to the previously obtained MsHPPD expression levels, in which lines OE2 and OE7 had the most significantly enhanced MsHPPD expression, compared with OE6 and wild-type (Fig. 4A). In Arabidopsis leaves, δ-tocopherol, γ-tocopherol, β-tocotrienol and γ-tocotrienol were not detected, and no significant difference was ultimately observed with regards to total vitamin E content or the levels of each quantifiable vitamin E form (Fig. S2B).
Figure 5. Analyses of vitamin E component in transgenic Arabidopsis seeds.
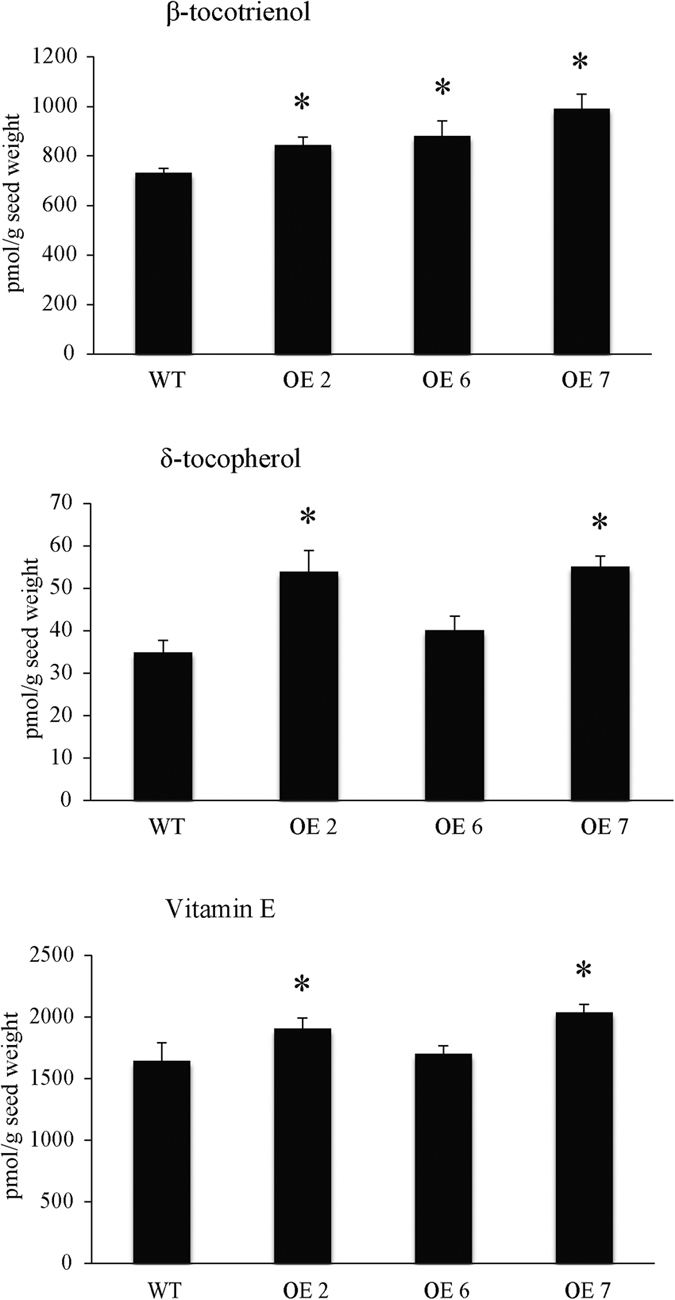
Seeds from MsHPPD-overexpressing lines (OE2, OE6 and OE7) and wild-type Arabidopsis were collected at the same time, air dried for two weeks and used for this measurement. Data are presented as mean ± SD with four biological replicates. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05.
Effect of MsHPPD on seed germination in Arabidopsis
To explore the potential role of the increased β-tocotrienol in transgenic Arabidopsis seeds, seed germination rate was examined under standard conditions, as well as under various stress conditions. Under normal conditions, the seed germination rate of MsHPPD-overexpressing transgenic lines was significantly higher than that of wild-type at 24 h. However, as the time after imbibition increased, the difference between the germination rates of transgenic and wild-type lines diminished, and so the difference in germination rate was less pronounced after 36 h, and by 48 h, both wild-type and transgenic lines reached equivalent germination rates under standard growth conditions (Fig. 6A). Under NaCl treatment, the seed germination rate of both transgenic and wild-type Arabidopsis lines was inhibited, however, the seed germination rate was significantly higher in the seeds of all transgenic lines compared to the corresponding rate observed in wild-type seeds, and this trend holds up at all tested time points. Ultimately, after 84 h, the germination rate of both the transgenic and control lines was determined to be 99% and 88%, respectively (Fig. 6B). Under 5 μM ABA treatment, the seed germination rate of MsHPPD-overexpressing transgenic lines at 120 h was approximately 86%, and this was significantly higher than the germination rate of the control line, which was found to be 40% (Fig. 6C). Furthermore, when the ABA concentration was doubled to 10 μM, the seed germination rate of the transgenic lines at 120 h decreased slightly to around 72%, but the germination rate of the wild-type lines decreased much more drastically to 18% (Fig. 6D).
Figure 6. Effect of MsHPPD on seed germination in transgenic Arabidopsis.
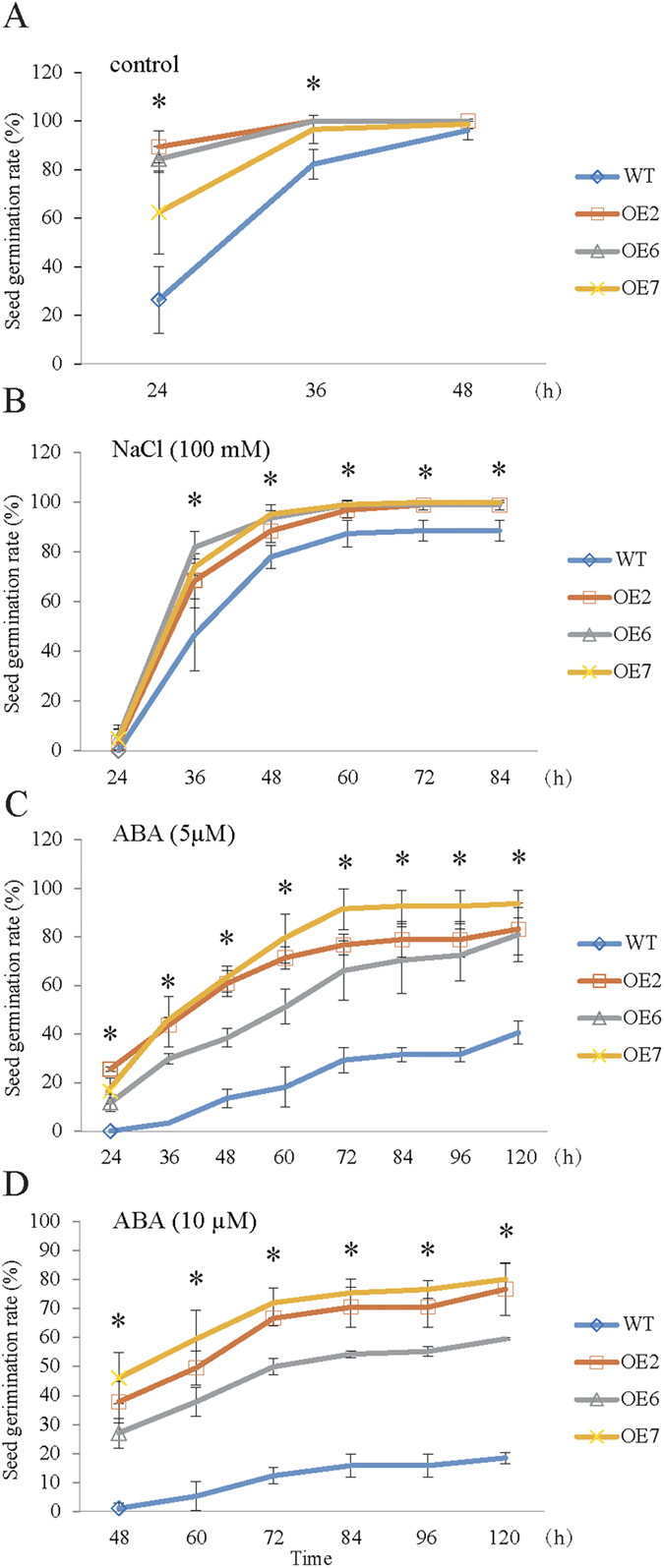
(A) Seed germination rate under normal conditions. (B) Seed germination rate under NaCl treatment. (C and D) Seed germination rate under ABA treatment. Seeds of transgenic and control Arabidopsis were harvested at the same time, dried at room temperature for two weeks, and germinated on half-strength MS medium, either with or without the applicable treatments. Seed germination rate was calculated based on four biological replicates (~80 seeds per replicate). Data are presented as mean ± SD. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05.
Expression of genes involved in the ABA biosynthetic and signaling and total ABA content are significantly reduced in seeds of MsHPPD-overexpressing Arabidopsis
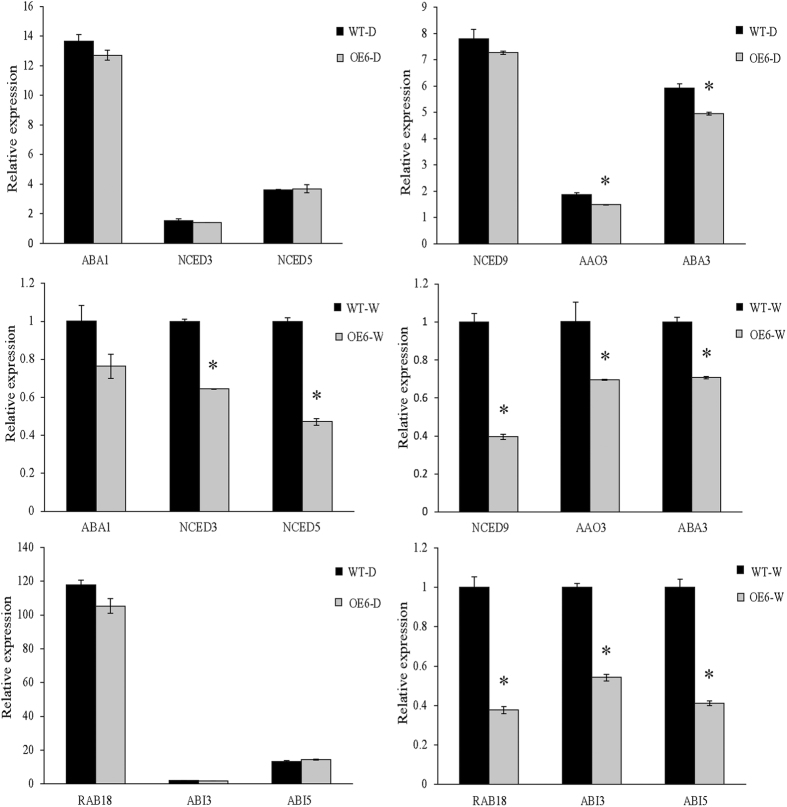
Since seed germination rate was significantly affected in the MsHPPD-overexpressing lines and ABA is an important phytohormone involved in seed dormancy and germination, we quantified the free ABA level in dry seeds from MsHPPD-overexpressing lines and wild-type. In addition, since a significant difference was observed in the seed germination rate between MsHPPD-overexpressing lines and wild-type imbibed seeds at 36 h, the ABA level in imbibed seeds were also measured. While no significant difference was observed in dry seeds, ABA content was significantly decreased in imbibed MsHPPD-overexpressing Arabidopsis seeds compared to imbibed wild-type seeds (Fig. 7A). Meanwhile, the expression levels of MsHPPD in MsHPPD-overexpressing lines were significantly higher in imbibed seeds than they were in dry seeds (Fig. 7B). Among the three MsHPPD-overexpressing lines, OE6 showed the dramatically highest MsHPPD expression level when imbibed with water and significantly reduced ABA content providing us with a suitable transgenic line to evaluate the relationship between MsHPPD expression level and ABA content. To determine whether the decrease of ABA levels in transgenic lines was due to changes in ABA biosynthesis or in ABA metabolism, qRT-PCR was performed to examine the expression levels of the ABA biosynthetic and signaling pathway genes in OE6. The results showed that the expression of the ABA biosynthetic genes ABA1, NCED3, NCED5 and NCED9 was not altered in dry seeds of the MsHPPD-overexpressing lines; however, in imbibed seeds, NCED3, NCED5 and NCED9 expression was significantly reduced, compared to the expression in wild-type seeds. Meanwhile, the expression levels of two other ABA biosynthetic genes, AAO3 and ABA3, were both significantly lower in the MsHPPD-overexpression lines than they were in wild-type seeds, regardless of whether the seeds were dried or imbibed. Likewise, the expression levels of the ABA signaling pathway genes RAB18, ABI3, and ABI5 were not changed in dry seeds, but were significantly down-regulated in imbibed seeds of the MsHPPD-overexpressing lines compared to wild-type seeds (Fig. 8).
Figure 7. Effects of MsHPPD on ABA content in transgenic Arabidopsis.
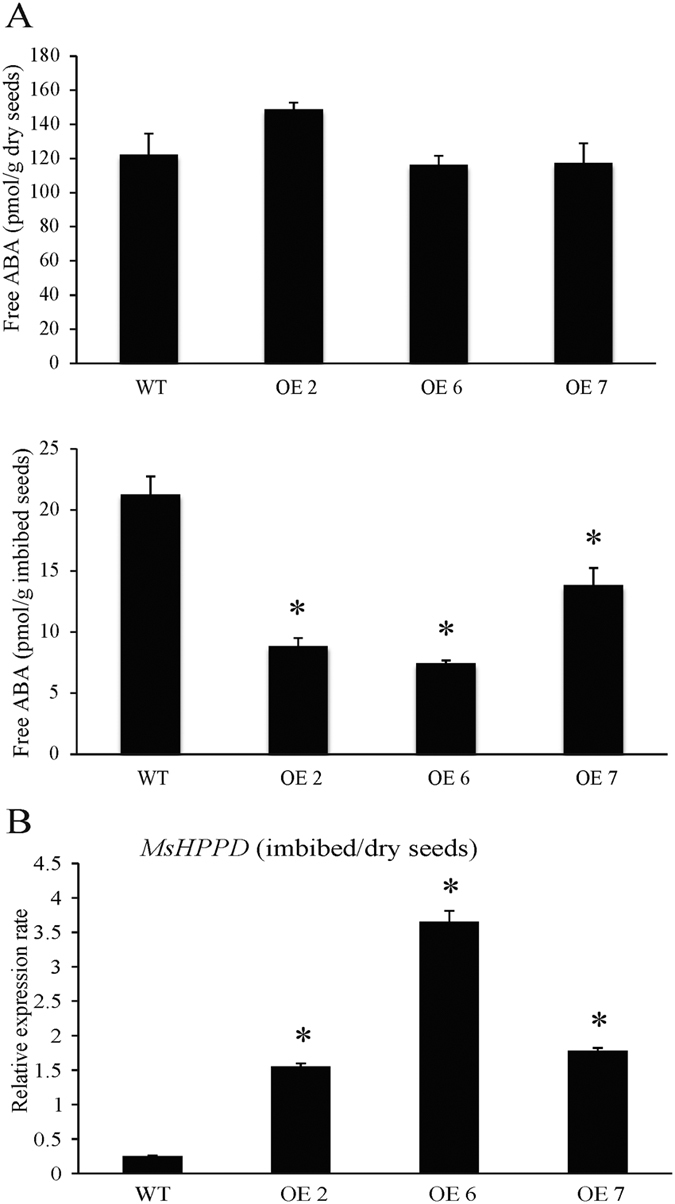
(A) ABA content in dry and imbibed MsHPPD-overexpressing Arabidopsis (OE2, OE6, and OE7) and wild-type seeds. Transgenic and control Arabidopsis seeds were harvested at the same time, dried at room temperature for two weeks, and used for ABA measurement in dry seeds (upper plot). Alternatively, they were submerged in water for 36 h and used for ABA measurement in imbibed seeds (lower plot). Data are presented as mean ± SD using three biological replicates. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05. (B) Expression rate of MsHPPD in imbibed seeds relative to dry seeds of MsHPPD-overexpressing transgenic and control Arabidopsis. The samples used here were the same as those in A. Data are presented as mean ± SD using three biological replicates and three technical replicates. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05.
Figure 8. Expression levels of ABA biosynthesis and ABA signaling pathway genes in MsHPPD-overexpressing transgenic Arabidopsis.
The samples used here were the same as those in Fig. 7. Data are presented as mean ± SD using three biological replicates and three technical replicates. The expression levels of the genes detected in the experiment were normalized to At4g26410. Statistical analyses were carried out via a two-tailed Student’s t-test with a significance of P < 0.05.
Discussion
HPPD catalyzes the first committed step in vitamin E biosynthesis2. Although it has been studied in many organisms27,28,29,30; the function of the HPPD gene in the economically important forage plant alfalfa remains unknown. In this study, full-length HPPD cDNA from alfalfa was isolated, and subsequent amino acid sequence alignment showed that MsHPPD from alfalfa shared high similarity with LsHPPD from Lactuca sativa, MtHPPD from Medicago truncatula, and AtHPPD from Arabidopsis thaliana. Among these HPPDs, LsHPPD and AtHPPD have been previously implicated as being able to alter vitamin E content in transgenic plants29,31. Phylogenetic tree construction also revealed that MsHPPD was evolutionally closest to HPPD sequences from these dicotyledonous plants.
Expression pattern analyses showed that NaCl and PEG treatment rapidly and robustly induced MsHPPD expression, suggesting that MsHPPD is involved in multiple stress responses in alfalfa. Darkness was also found to induce the expression of MsHPPD, and this is in agreement with the results from Falk et al.32, who found that HPPD expression levels increased during dark-induced leaf senescence. This may be due to the breakdown of chlorophyll-released free phytol, a precursor for tocopherol synthesis, and thus increased the demand for HPPD33,34. When plants were transferred from darkness to light, MsHPPD expression dropped sharply and continued to decrease (Fig. 3D), which suggests that light acts as a negative regulator of MsHPPD expression.
ABA induced the expression of MsHPPD modestly, albeit significantly. MsHPPD expression levels were initially high in primary roots and cotyledons, and increased significantly after the ABA induction. It has been shown that salicylic acid (SA), methyl jasmonic acid (MeJA), and high light can induce the expression of both SmHPPD from Salvia miltiorrhiza and also AtHPPD30,35. MeJA treatment was not found to alter the expression levels of MsHPPD, which might be a manifestation of the lack of a MeJA-responsive element in the promoter region of MsHPPD (Fig. S1). However, in agreement with the previous study, SA has significantly induced the expression of MsHPPD. Taken together, our results suggest that HPPD could be induced by multiple conditions, and the flexible expression levels of MsHPPD in response to stresses implied that MsHPPD plays multiple important roles with regards to responding to stresses.
In the leaves of MsHPPD-overexpressing transgenic lines, no difference was recorded to either vitamin E composition or vitamin E content (Fig. S2). This was contradictory to the result obtained by Tsegaye et al.31, who reported that overexpressing AtHPPD increased tocopherol by around 37% in transgenic Arabidopsis leaves. However, overexpressing barley HPPD had no effect on the tocopherol content in transgenic tobacco leaves11. This discrepancy may be attributable to subtle differences between the different organisms used in these studies. To gain a deeper insight into the reason for this discrepancy, we tested the expression levels of endogenous vitamin E biosynthetic genes in Arabidopsis to examine whether MsHPPD overexpression affected the flux of the entire vitamin E biosynthetic pathway. Interestingly, we observed a significant decrease only in the expression level of endogenous AtHPT in transgenic lines, suggesting that the vitamin E biosynthesis pathway is tightly controlled. The increased expression levels of MsHPPD appear to have somehow inhibited the expression of AtHPT, which also plays a role in determining vitamin E accumulation along with HPPD, and the inhibition of AtHPT may counteract the effects caused by the enhancement of MsHPPD expression on vitamin E to an unknown degree. As such, perhaps this explains why no significant differences were observed regarding the vitamin E content of the leaves.
As was previously reported, the content and composition of vitamin E varies in different tissues and different developmental stages8. α-tocopherol is the dominant form found in plant leaves, while γ-tocopherol and tocotrienols mainly exist in seeds36. This phenomenon made us question the function of elevated β-tocotrienol in seeds; therefore, we tested the seed germination rate of seeds from MsHPPD-overexpressing Arabidopsis lines and also wild-type seeds, under both normal and stressful conditions. Under normal conditions, the transgenic lines germinated earlier than wild-type plants. Meanwhile, under NaCl treatment, the germination of both transgenic and wild type lines was inhibited compared to normal conditions; however, the germination rate of the seeds from transgenic lines was significantly higher than that of wild type seeds. These results suggest that MsHPPD plays a positive role in regulating seed germination.
Seed eFP browser37 showed that HPPD belongs to the ABA group of genes and that it might be involved in ABA biosynthesis or the ABA signaling pathway. Here, we showed that transgenic Arabidopsis lines which overexpressing MsHPPD were less sensitive to ABA treatment, in terms of seed germination rate (Fig. 6C,D). Additionally, the ABA content was significantly lower in imbibed transgenic Arabidopsis seeds than it was in imbibed wild-type seeds. However, the expression level of MsHPPD was significantly higher in imbibed transgenic Arabidopsis seeds. Since vitamin E and ABA are both downstream of GGDP, overexpression of MsHPPD could increase in vitamin E content, thus decreasing the amount of GGDP substrate available for ABA biosynthesis. Expression of the ABA biosynthetic genes NCED3, NCED5 and NCED9 was found to be unaltered in dry seeds; but was significantly reduced in imbibed transgenic Arabidopsis seeds (Fig. 8), suggesting that the reduced GGDP pool size decreased the expression level of ABA biosynthetic genes. In addition, expression of the ABA signaling pathway genes ABI3 and ABI5, which are important for seed germination, decreased significantly in imbibed transgenic Arabidopsis seeds. Based on the above data, it is possible that vitamin E and ABA compete for GGDP, and, thus, elevated vitamin E necessitate reduced ABA levels, and indeed, this is what we observed (Fig. S3). In turn, the reduced ABA content led to a weakening of ABA signaling, ultimately resulting in either a faster-than-normal or higher-than-normal germination rate in the seeds of the transgenic MsHPPD-overexpressing lines, compared with the germination rate of the corresponding control, under various conditions. Moreover, vitamin E has been demonstrated to have additional biological activities besides its antioxidant functions in mammalian systems. Specific tocopherols have been found to modulate signal transduction pathways38,39. For instance, tocopherol-binding proteins can regulate gene transcriptional activities40. Although a regulatory activity for tocopherols has not yet been directly proven in land plants, the present data raises the possibility that vitamin E also modulates signal transduction pathways in plants. Specifically, tocopherol may modulate seed germination and other hormone responses by binding with other proteins to regulate the ABA signaling pathway. Additional work is necessary for these hypotheses to be investigated in future studies.
In conclusion, an HPPD gene that encodes a 434 amino acid protein, named MsHPPD, was isolated from alfalfa. MsHPPD was shown to be induced by multiple treatments, and overexpression of MHPPD in Arabidopsis increased the total vitamin E content. Seed germination of the transgenic lines was accelerated under normal conditions, and was less strongly inhibited under stress treatments. Overexpression of MsHPPD was also found to significantly reduce the free ABA content, as well as the expression of ABA biosynthetic genes and ABA signaling pathway genes. This represents the first evidence that vitamin E is involved in regulating germination rate via affecting the ABA biosynthesis and the subsequent ABA signaling pathway.
Methods
Plant materials and growth conditions
Arabidopsis (Columbia-0) was used for these present experiments. Seeds were surface sterilized with 70% ethanol for 2 min and 5% sodium hypochlorite for 5 min, and then washed three times with water. The sterilized seeds were planted on half-strength MS medium (2.2 g/L Murashige and Skoog salts, pH 5.7, and 8 g/L agar), and kept at 4 °C for 2 days to stratify the seeds and facilitate seed germination. The Arabidopsis plants were grown under long-day photoperiod conditions (16 h light/8 h dark at 25 °C). Seeds from Medicago sativa L. cv Zhongmu No. 1 were germinated on wet filter paper, and the seedlings were grown under long-day photoperiod conditions as above, after the appearance of the first true leaf.
Gene cloning, vector construction and transformation
The full-length MsHPPD gene was cloned from cDNA prepared from alfalfa leaves via rapid amplification of cDNA ends (RACE), according to the manufacturer’s protocol (Clontech, USA). The promoter of MsHPPD was cloned via chromosome walking. The binary vector pBI12141 with a GUS reporter gene was used to express MsHPPD under the control of the constitutive CaMV 35 S promoter, and the CDS of MsHPPD without a stop codon was introduced into PBI121 and fused with the GUS reporter gene. For the gene expression analysis, the CaMV 35 S promoter was replaced with the native promoter of MsHPPD, which was fused to the GUS reporter gene. The constructs harboring MsHPPD and pMsHPPD were transformed into the agrobacterial strain GV3101, which was then used for Arabidopsis transformation via the well-established floral dipping method42.
Treatments
Three-week-old Zhongmu No. 1 seedlings, which were grown in half-strength MS nutrient water, were subjected to different treatments. NaCl, ABA, PEG, SA, and MeJA treatments were performed by adding the compounds to the nutrient water and collecting leaf tissue at 0, 2, 4, 8, 12 and 24 h after treatment. The final concentrations of NaCl, ABA, PEG, SA, and MeJA were 100 mM, 0.1 mM, 300 mM, 0.1 mM, and 0.1 mM, respectively43. Light treatment was performed by growing seedlings in dark conditions, and then collecting leaf tissue at 0, 12, 24, 48, and 72 h, and a second group of experiments was performed by exposing 72 h dark-treated seedlings to light and collecting leaf tissue at 0.25, 0.5, 6, and 12 h.
Quantitative real-time PCR
Total RNA was isolated using TRIzol (Invitrogen, USA) and treated with DNase I to eliminate DNA contamination. One microgram of total RNA was then used for cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen, USA). qRT-PCR was performed with an ABI 7500 Real-Time PCR System (Bio-Rad, USA) using the SYBR Green PCR Master Mix (Takara, Japan) and the appropriate primers (Table 1). The relative gene expression levels were calculated by 2−ΔΔCt 44. At4g2641045 and Actin25 were used as reference genes to normalize the expression levels of the target genes in Arabidopsis and alfalfa, respectively. All experiments were performed using three biological replicates and three technical replicates.
Table 1. The primer sequences used in cloning and expression analysis.
| Primers | Sequences | Application |
|---|---|---|
| 3′RACE | GTGCCATTCCATCGTCTCCTCCA | RACE |
| 5′RACE | GGATTCTGGTTTGGGTTCGGGTC | RACE |
| PBI-HPPD-F | CACGGGGGACTCTAGAATGGCCATCGAAACAGAAAC | Vector construction |
| PBI-HPPD-R | AGGGACTGACCACCCGGGGATGCAGTTCTTCTAGTTTCCA | Vector construction |
| AtHPPD-F | GCGGTTTAAATTCAGCGGTCCTG | Q-PCR |
| AtHPPD-R | TCCGTGCACTGGCTCGTTAATC | Q-PCR |
| AtHPT-F | TGGAGCAAAGTCATCTCGGTTG | Q-PCR |
| AtHPT-R | GACTTAGCTCGAGCCCACAAAG | Q-PCR |
| AtTC-F | TGTCCGAAGGGTTCCAAGCTAC | Q-PCR |
| AtTC-R | CACAGTTTCCGCATAGTCAGTACG | Q-PCR |
| AtMPBQMT-F | TGCCGGTTTCAAGGACGTTCAG | Q-PCR |
| AtMPBQMT-R | TCTTCCTTTGGACCAAGCTGGAG | Q-PCR |
| AtTMT-F | TGCTCAATCACTCGCTCATAAGGC | Q-PCR |
| AtTMT-R | TCTTCGAATGGCTGATCCAACGC | Q-PCR |
| VTE5-F | CAAGCGAAACGTCATTCAACAGAG | Q-PCR |
| VTE5-R | GAAGATTGGCCACGCAAGTACG | Q-PCR |
| AtHST-F | GCATGTTCTCAGGTTGGTGCTG | Q-PCR |
| AtHST-R | AGCTCTTGTCACCAAGGCAGTG | Q-PCR |
| AtABI3-F | GAAGCAAAGCGACGTGGGTAAC | Q-PCR |
| AtABI3-R | AACCTGTAGCGCATGTTCCAAAC | Q-PCR |
| AtABI5-F | AAACATGCATTGGCGGAGTTGG | Q-PCR |
| AtABI5-R | CGGTTGTGCCCTTGACTTCAAAC | Q-PCR |
| AtABA1-F | TGGGCTTGGTCCTCTGTCTTTC | Q-PCR |
| AtABA1-R | CACCAACTCTTCCTGGATGTGG | Q-PCR |
| AtRAB18-F | TCTAGCTCGGAGGATGATGGAC | Q-PCR |
| AtRAB18-R | ACCGTAGCCACCAGCATCATATC | Q-PCR |
| AtNCED3-F | TCGAAGCAGGGATGGTCAACAG | Q-PCR |
| AtNCED3-R | GCTCGGCTAAAGCCAAGTAAGC | Q-PCR |
| AtNCED5-F | CCTCCGTTAGTTTCACCAACACT | Q-PCR |
| AtNCED5-R | GGTGTGTCGGAGACGGAGTT | Q-PCR |
| AtNCED9-F | CCCAAACCGCAGCGTTTAAT | Q-PCR |
| AtNCED9-R | GTTCGGTCGAGGAATTGGGT | Q-PCR |
| AtABA3-F | TGCTACAAGGCTCCCCCTTT | Q-PCR |
| AtABA3-R | CACAACCCTTTGCAGCATCA | Q-PCR |
| AtAAO3-F | GAAGGTCTTGGAAACACGAAGAA | Q-PCR |
| AtAAO3-R | GAAATACACATCCCTGGTGTACAAAAC | Q-PCR |
| MsHPPD-F | CAGTTATCCTCGAAAACCACG | Q-PCR |
| MsHPPD-R | GTTTGGGTTCGGGTCGTTG | Q-PCR |
| Actin-F | GAGCGTTTCCGTTGTCCTGA | Q-PCR |
| Actin-R | AGGTGCTGAGGGAAGCCAAA | Q-PCR |
GUS histochemical assay
A GUS histochemical assay was performed according to the method described by Jefferson et al.46. Plant tissues were incubated at 37 °C overnight in 50 mM sodium pyrophosphate staining buffer (pH 7.2) containing 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl β-D-glucuronide), 10 mM EDTA, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 0.2% (v/v) Triton X-100. 100% ethanol was used to completely remove chlorophyll from tissues.
Extraction and HPLC analyses of tocopherols and tocotrienols
Extractions and analyses of tocopherols and tocotrienols were performed according to the method described before25.
Seed germination
MsHPPD-overexpressing and wild-type Arabidopsis seeds were surface-sterilized as described above and planted on half-strength MS medium with the presence or absence of 100 mM NaCl, 5 μM ABA, or 10 μM ABA. After being stratified at 4 °C for 3 days, the seeds were transferred to a growth chamber (16 h light/8 h dark at 25 °C) and germination rate was monitored every 12 hours.
ABA measurement in Arabidopsis seeds
For each replicate, approximately 20 mg (dry weight) of seeds were homogenized under liquid nitrogen, weighed, and extracted for 24 h in cold methanol and 2H6-ABA (internal standard, OlChemIm, Czech Republic). Endogenous ABA was purified and measured as previously described47 with some modifications to the detection conditions. LC-MS/MS analysis was performed on a UPLC system (Waters) coupled to the 5500 QTRAP system (AB SCIEX, USA), and a BEH C18 column (1.7 mm, 2.1 × 150 mm; Waters) was used for LC separation with mobile phase 0.05% HAc (A) and 0.05% HAc in ACN (B). The gradient was set with initial 20% B and increased to 70% B within 6 min. ABA was detected in MRM mode with transition 263/153. Finally, quantitation was performed using the isotope dilution method.
Statistical analysis
Statistical analyses were performed by two-tailed Student’s t-test. The difference was considered significant at P < 0.05. Data presented are mean ± SD.
Additional Information
How to cite this article: Jiang, J. et al. P-HYDROXYPHENYLPYRUVATE DIOXYGENASE from Medicago sativa is involved in vitamin E biosynthesis and abscisic acid-mediated seed germination. Sci. Rep. 7, 40625; doi: 10.1038/srep40625 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank Cade Guthrie, Mark Lemos (University of California, Davis) and Yongzhen Pang (Chinese Academy of Sciences) for their critical reading of the article. This work was supported by grants from the National Basic Research Program of China (2014CB138703), National Program for Forage Germplasm Conservation (2130135) and the Agricultural Science and Technology Innovation Program (ASTIP-IAS10).
Footnotes
Author Contributions J.J. and X.W. designed the experiments and wrote the manuscript; J.J. and J.H. performed the experiments and analyzed the data. H.G. supervised the study; Z.C. and L.B. proofread the language of the manuscript; Y.W. carried out the GUS histochemical stain assay; J.C. and S.F. performed the free ABA measurement, Z.W. cultured the plants which were used in this research.
References
- Miret J. A. & Munne-Bosch S. Redox signaling and stress tolerance in plants: a focus on vitamin E. Ann N Y Acad Sci. 1340, 29–38 (2015). [DOI] [PubMed] [Google Scholar]
- Abbasi A. R., Hajirezaei M., Hofius D., Sonnewald U. & Voll L. M. Specific roles of alpha- and gamma-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol. 143, 1720–1738 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco A., Mattioli S., Ruggeri S., Mugnai C. & Castellini C. Effect of slaughtering age in different commercial chicken genotypes reared according to the organic system: 2. Fatty acid and oxidative status of meat. Ital J of Anim Sci. 13 (2014). [Google Scholar]
- Traber M. G. Vitamin E inadequacy in humans: causes and consequences. Adv Nutr. 5, 503–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H. M. & Bishop K. S. On the Existence of a Hitherto Unrecognized Dietary Factor Essential for Reproduction. Science. 56, 650–651 (1922). [DOI] [PubMed] [Google Scholar]
- Sattler S. E. Characterization of Tocopherol Cyclases from Higher Plants and Cyanobacteria. Evolutionary Implications for Tocopherol Synthesis and Function. Plant Physiol. 132, 2184–2195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D. & Sonnewald U. Vitamin E biosynthesis: biochemistry meets cell biology. Trends Plant Sci. 8, 6–8 (2003). [DOI] [PubMed] [Google Scholar]
- DellaPenna D. A decade of progress in understanding vitamin E synthesis in plants. J Plant Physiol. 162, 729–737 (2005). [DOI] [PubMed] [Google Scholar]
- Sakuragi Y., Maeda H., Dellapenna D. & Bryant D. A. Alpha-Tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141, 508–521 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S. R., Barrette T. R. & DellaPenna D. Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 7, 2139–2149 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J., Andersen G., Kernebeck B. & Krupinska K. Constitutive overexpression of barley 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Letters. 540, 35–40 (2003). [DOI] [PubMed] [Google Scholar]
- Karunanandaa B. et al. Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab Eng. 7, 384–400 (2005). [DOI] [PubMed] [Google Scholar]
- Crowell E. F., McGrath J. M. & Douches D. S. Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Res. 17, 205–217 (2008). [DOI] [PubMed] [Google Scholar]
- Seiler C. et al. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot. 62, 2615–2632 (2011). [DOI] [PubMed] [Google Scholar]
- Guajardo E., Correa J. A. & Contreras-Porcia L. Role of abscisic acid (ABA) in activating antioxidant tolerance responses to desiccation stress in intertidal seaweed species. Planta. 243, 767–781 (2015). [DOI] [PubMed] [Google Scholar]
- Nambara E. & Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 56, 165–185 (2005). [DOI] [PubMed] [Google Scholar]
- Hirai N., Yoshida R., Todoroki Y. & Ohigashi H. Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci Biotechnol Biochem. 64, 1448–1458 (2000). [DOI] [PubMed] [Google Scholar]
- DellaPenna D. & Pogson B. J. Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol. 57, 711–738 (2006). [DOI] [PubMed] [Google Scholar]
- Lefebvre V. et al. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319 (2006). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. ABA-HYPERSENSITIVE BTB/POZ PROTEIN 1 functions as a negative regulator in ABA-mediated inhibition of germination in Arabidopsis. Plant Mol. Biol. 90, 303–315 (2016). [DOI] [PubMed] [Google Scholar]
- Komatsu K. et al. Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun. 4, 2219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke J. A. et al. The Medicago sativa gene index 1.2: a web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genomics. 16, 502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. et al. Isolation and functional characterization of salt-stress induced RCI2-like genes from Medicago sativa and Medicago truncatula. J Plant Res. 128, 697–707 (2015). [DOI] [PubMed] [Google Scholar]
- Armstrong J. M. CYTOLOGICAL STUDIES IN ALFALFA POLYPLOIDS. Canadian Journal of Botany. 32, 531–542 (1954). [Google Scholar]
- Jiang J. et al. Overexpression of Medicago sativa TMT elevates the alpha-tocopherol content in Arabidopsis seeds, alfalfa leaves, and delays dark-induced leaf senescence. Plant Sci. 249, 93–104 (2016). [DOI] [PubMed] [Google Scholar]
- Lescot M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matringe M., Ksas B., Rey P. & Havaux M. Tocotrienols, the unsaturated forms of vitamin E, can function as antioxidants and lipid protectors in tobacco leaves. Plant Physiol. 147, 764–778 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S. et al. Simultaneous expression of Arabidopsis p-hydroxyphenylpyruvate dioxygenase and MPBQ methyltransferase in transgenic corn kernels triples the tocopherol content. Transgenic Res. 20, 177–181 (2011). [DOI] [PubMed] [Google Scholar]
- Ren W. et al. Molecular cloning and characterization of 4-hydroxyphenylpyruvate dioxygenase gene from Lactuca sativa. J Plant Physiol. 168, 1076–1083 (2011). [DOI] [PubMed] [Google Scholar]
- Xiao Y. et al. Characterization and expression profiling of 4-hydroxyphenylpyruvate dioxygenase gene (Smhppd) from Salvia miltiorrhiza hairy root cultures. Mol Biol Rep. 36, 2019–2029 (2009). [DOI] [PubMed] [Google Scholar]
- Tsegaye Y., Shintani D. & DellaPenna D. Overexpression of the enzyme p-hydroxyphenolpyruvate dioxygenase in arabidopsis and its relation to tocopherol biosynthesis. Plant Physiol. Biochem. 40, 913–920 (2002). [Google Scholar]
- Chrost B., Falk J., Kernebeck B., Mölleken H. & Krupinska K. In The Chloroplast: From Molecular Biology to Biotechnology (eds Argyroudi-Akoyunoglou Joan H. & Senger Horst) 171–176 (Springer, Netherlands, 1999). [Google Scholar]
- Valentin H. E. et al. The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell. 18, 212–224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Dorp K. et al. Remobilization of Phytol from Chlorophyll Degradation Is Essential for Tocopherol Synthesis and Growth of Arabidopsis. Plant Cell. 27, 2846–2859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E. & DellaPenna D. The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol. 133, 930–940 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon E. B. et al. Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol. 21, 1082–1087 (2003). [DOI] [PubMed] [Google Scholar]
- Bassel G. W. et al. Elucidating the germination transcriptional program using small molecules. Plant Physiol. 147, 143–155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen C. K., Khanna S., Roy S. & Packer L. Molecular basis of vitamin E action. Tocotrienol potently inhibits glutamate-induced pp60(c-Src) kinase activation and death of HT4 neuronal cells. J Biol Chem. 275, 13049–13055 (2000). [DOI] [PubMed] [Google Scholar]
- Nobata Y. et al. Alpha-Tocopherol Inhibits IL-8 synthesis induced by thrombin and high glucose in endothelial cells. Horm Metab Res. 34, 49–54 (2002). [DOI] [PubMed] [Google Scholar]
- Yamauchi J. et al. Tocopherol-associated protein is a ligand-dependent transcriptional activator. Biochem Biophys Res Commun. 285, 295–299 (2001). [DOI] [PubMed] [Google Scholar]
- Chen P.-Y., Wang C.-K., Soong S.-C. & To K.-Y. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Molecular Breeding. 11, 287–293 (2003). [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Chen T. et al. Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol Biol Rep. 39, 6067–6075 (2012). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Walley J. W. et al. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4, e1000237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A. & Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J. & Yan C. Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci. 28, 1081–1087 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.