Abstract
With the increase in sea surface temperature (SST), scleractinian corals are exposed to bleaching threats but may possess certain flexibilities in terms of their associations with symbiotic algae. Previous studies have shown a close symbiosis between coral the and Symbiodinium; however, the spatial variation of the symbiosis and the attribution underlying are not well understood. In the present study, we examined coral-algal symbiosis in Galaxea fascicularis and Montipora spp. from three biogeographic regions across ~10° of latitude in the South China Sea. Analysis of similarities (ANOSIM) indicated a highly flexible coral-algal symbiosis in both G. fascicularis and Montipora spp. and canonical correspondence analysis (CCA) showed that temperature explained 83.2% and 60.1% of the explanatory subclade variations in G. fascicularis and Montipora spp., respectively, which suggested that temperature was the main environmental factor contributing to the diversity of Symbiodinium across the three regions. The geographic specificity of the Symbiodinium phylogeny was identified, revealing possible environmental selection across the three regions. These results suggest that scleractinian corals may have the ability to regulate Symbiodinium community structures under different temperatures and thus be able to adapt to gradual climate change.
Under global climate change, coral reefs are greatly threatened by reduced productivity and the precipitation of calcium carbonate1,2. The symbiotic dinoflagellates (Symbiodinium) in corals contribute approximately 95% of the coral energy requirements3 and play a central role in coral reef maintenance and productivity. In addition, coral reef resilience to future climate change strongly relies on the adaptation of coral-algal symbiosis. According to previous studies, a continuous loss of Symbiodinium, such as a bleaching event, dramatically increases coral mortality. Hence, an understanding of coral-algal symbiosis change in response to different environmental conditions is essential to develop coral reef conservation strategies under future climate change.
The genus Symbiodinium consists of nine clades (i.e., clades A–I) with varied photosynthetic efficiencies and heat tolerance abilities4. Although clades A–D are the main Symbiodinium harbored by scleractinian corals, some other clades, such as F/G, have also been found in scleractinian corals and have been proposed to exert certain functional roles in the coral reef ecosystem5. The flexibility of coral-algal symbiosis is widely accepted6,7,8. Because corals are able to change their Symbiodinium communities in response to diverse environmental stressors, for example, by gaining heat-tolerant Symbiodinium in an environment with an elevated temperature, the newly formed coral-algal symbiosis has been hypothesized to be more beneficial for coral survival9. In contrast, on a large latitude scale, some coral species still maintain a very stable coral-algal symbiosis10. Thus, the capacity for coral-algal symbiosis change in response to environmental change remains controversial.
Temperature has been shown to have a great impact on coral-algal symbiosis11,12. At extremely high temperatures, Symbiodinium might be expelled from the coral host. After recovering from bleaching or remaining a sufficient duration in a relatively warm temperature, the coral host can gain some heat-tolerant Symbiodinium9. In addition, nutrients, light and salinity all have been shown to affect Symbiodinium community structures13,14,15; however, among these environmental factors, the one(s) with the greatest effect remains unclear.
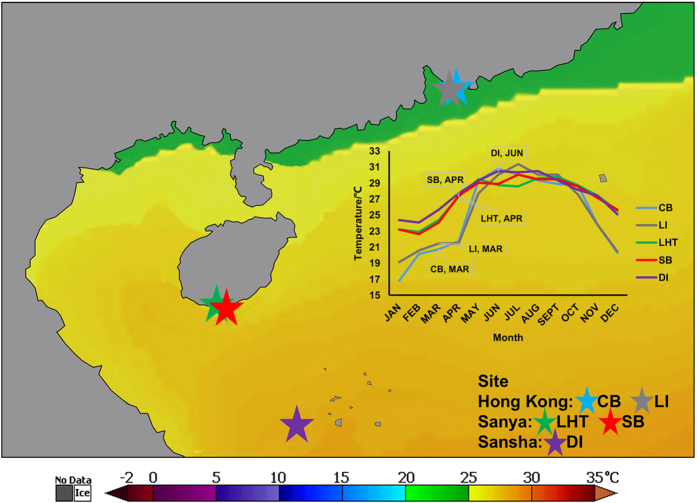
Hong Kong (Table 1) is located at 22°10′N to 22°30′N and has a subtropical climate with low levels of salinity in the summer16, in which the sea surface temperature (SST) ranges from 13 to 30 °C which is marginal for hard coral growth17. Because of its extreme seasonal temperature changes and relatively low salinity, there are mainly northern marginal corals distributed in Hong Kong. Sanya and Sansha, lying in the southern region of Hainan Province, have a tropical climate with SST ranges from 20 to 30 °C and from 24 to 30 °C, respectively. Coral reefs in Sanya are intermediate and are located near the northern border of the global distribution of tropical coral reefs18, whereas coral reefs in Sansha are typically tropical. In general, the average annual SST rises across the three regions with changes in latitude (Fig. 1). The impact of human activities on the five sampling sites is as follows: Lamma Island (LI) > Crescent Bay (CB), Luhuitou (LHT) > Sunny Bay (SB) > Drummond Island (DI). Crescent Bay is located in the northeast of Hong Kong, where there is marginal influence by the Pearl River and the greatest coral cover (30–50%) in Hong Kong has been observed. In contrast, Lamma Island in the southwest of Hong Kong, has a relatively low coral cover (10–30%) due to the influence of the Pearl River16. The coral cover in Luhuitou of Sanya decreased from 80–90% in the 1960 s to 11% in 2007 due to frequent human activities; the coral cover around Sunny Bay in Sanya was approximately 35% in the period from 2007–200919. Drummond Island in the Sansha region is one of the Xisha Islands with a coral cover of approximately 50% based on 30-year data from the 1970 s20. We selected these five sampling sites with different environmental conditions to examine the change of coral-algal partnerships in response to environmental variations and to identify the environmental factor(s) with the greatest impact.
Table 1. Sampling sites, sampling dates and sample ID abbreviations for G. fascicularis and Montipora spp.
| Regions | Climate | Sites | Coordinates | Dates (yyyy-mm-dd) | G. fascicularis | Montipora spp. |
|---|---|---|---|---|---|---|
| Hong Kong | Subtropical | Crescent Bay (CB) | E114.314°, N22.531° | 2014-03-24 | G. fascicularis (GA) | M. peltiformis (MO) |
| Lamma Island (LI) | E114.135°, N22.187° | 2014-03-19 | N.A. | M. venosa (MO) | ||
| Sanya | Tropical | Luhuitou (LHT) | E109.471°, N18.212° | 2014-04-04 | G. fascicularis (GA) | M. monasteriata (MO) |
| Sunny Bay (SB) | E109.610°, N18.199° | 2014-04-03 | G. fascicularis (GA) | M. monasteriata (MO) | ||
| Sansha | Tropical | Drummond Island (DI) | E111.778°, N16.523° | 2014-06-19 | G. fascicularis (GA) | N.A. |
Figure 1. Sampling sites in the South China Sea: Crescent Bay (CB, Hong Kong), Lamma Island (LI, Hong Kong), Luhuitou (LHT, Sanya), Sunny Bay (SB, Sanya) and Drummond Island (DI, Sansha).
The plot displays mean SST for each sampling site in 2014 (NASA Earth Observations) and the labels indicate sampling month for each site. The temperature heatmap was generated on the NOAA Satellite and Information Service website (http://coralreefwatch.noaa.gov/satellite/bleaching5km/index_5km_sst.php) and using NOAA Coral Reef Watch Virtual Stations SST data for 2014 (NOAA Coral Reef Watch. 2014, updated daily. NOAA Coral Reef Watch Annual 5-km Satellite Sea Surface Temperature Product for Coral Triangle, Jan. 1, 2014–Dec. 31, 2014. College Park, Maryland, USA: NOAA Coral Reef Watch. Data set accessed at http://coralreefwatch.noaa.gov/satellite/bleaching5km/index_5km_sst.php) shows the thermal gradient in the South China Sea.
The diversity of Symbiodnium has mostly been examined by denaturing gradient gel electrophoresis (DGGE) based analysis, which is unable to detect Symbiodinium subclades with a relative abundance of less than 10% and thus potentially lacks the sufficient sensitivity to detect symbiosis change7. The development of highly sensitive molecular techniques, such as high-throughput DNA sequencing, would facilitate the identification of low abundance Symbiodinium that benefit coral hosts in terms of tolerant to climate change. Additionally, amplicon sequencing using ITS2 primers has been shown to be reliable for measurements of Symbiodinium communities10. In the present study, to explore spatial variations of coral-algal symbiosis, we used ITS2 amplicon sequencing to examine coral-algal symbiosis in G. fascicularis and Montipora spp. from five sampling sites across three biogeographic regions, including tropical and subtropical seas, and we attempted to elucidate the symbiosis change across these regions. Furthermore, we examined the effects of temperature, depth, salinity etc. on coral-algal symbiosis change.
Results
Sequence information
In total, 1,774,246 sequences (more than 80% of the input sequences) were retained for further analysis after quality control. After removing the fusion primers, the median length of the sequences was ~332 bp. The dataset included a total of 58 samples: six samples of G. fascicularis from CB(Hong Kong), LHT(Sanya), SB(Sanya) and DI(Sansha) each (CB_GA1-6, LHT_GA1-6, SB_GA1-6, DI_GA1-6), six samples of Montipora spp. from LI(Hong Kong) and SB(Sanya) each (LI_MO1-6, SB_MO1-6), four samples of Montipora sp. (LHT_MO1-4), two Montipora sp. technical replicates (LHT_MO5-6) from LHT(Sanya), five samples of Montipora sp. (CB_MO1-5), a Montipora sp. (CB_MO6) technical replicate from CB(Hong Kong) and two samples of seawater from each sampling site (CB_SW1-2, LI_SW1-2, LHT_SW1-2, SB_SW1-2, DI_SW1-2). Three Montipora spp. samples were failed during DNA extraction or PCR amplification. In total, 131 Symbiodinium subclades were assigned based on alignment to the ITS2 database according to 97% similarity (whole_community_data.xls, Supporting Information), and 13 dominant subclades (covering more than 85% of the sequence) were selected for further analyses according to the relative abundance of the Symbiodinium subclade, the rest were regarded as “Others”.
Disparity in Symbiodinium community structures of G. fascicularis and Montipora spp. across the three geographic regions
Clade A, clade B, clade C, clade D and clade F were detected in samples. Among all samples, only two seawater samples from Sansha contained 0.06% and 0.6% A clade, respectively. The total relative abundance of clade A, B and F in samples were all less than 0.01%, thus only Symbiodinium clade C and D were kept for further analysis. As shown in Fig. 2, G. fascicularis in Sanya and Sansha had a high symbiont specificity for clade D, whereas G. fascicularis in Hong Kong showed a high symbiont specificity for clade C. In contrast, in all sampling sites, Montipora spp. contained a very low abundance of clade D but a high abundance of clade C. All of the seawater samples were dominated by clade C, suggesting Symbiodinium selection and enrichment of G. fascicularis in Sanya and Sansha.
Figure 2. Symbiodinium community compositions at the clade level.
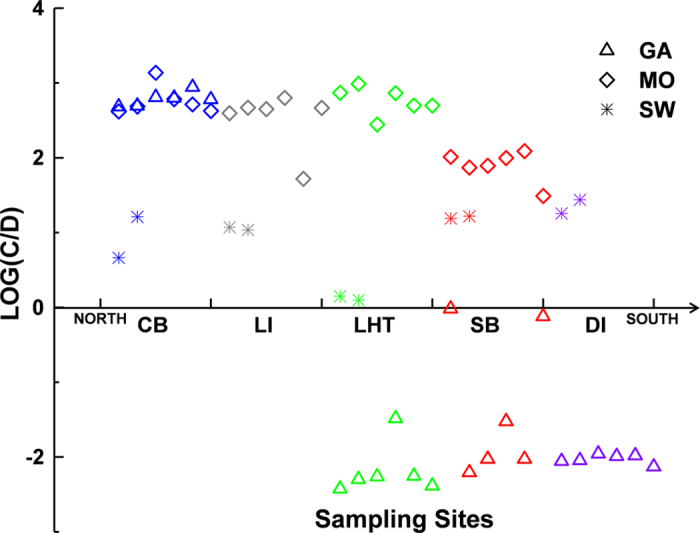
Compositions of dominant Symbiodinium clades from Crescent Bay (CB, Hong Kong), Lamma Island (LI, Hong Kong), Luhuitou (LHT, Sanya), Sunny Bay (SB, Sanya), Drummond Island (DI, Sansha) in the South China Sea. Triangles, diamonds, stars represent G. fascicularis (GA), Montipora spp. (MO), and seawater (SW) samples from each site, respectively. Each symbol represents a single sample and the value is calculated by logarithm of the ratio of C clade relative abundance to D clade relative abundance in each sample.
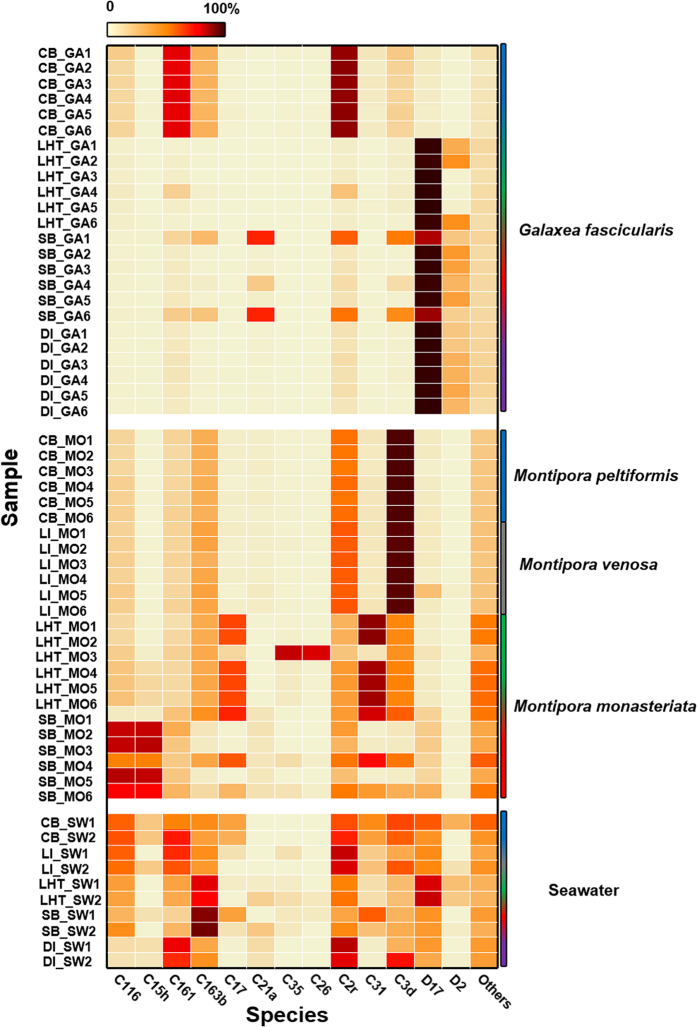
At the subclade level, 13 dominant/subdominant subclades were found, accounting for more than 85% of the total sequences (Fig. 3). These subclades were picked as their relative abundances were >10% in at least one sample or their average relative abundances in all samples were at least around 1%. In G. fascicularis, C161, C21a, C2r, D17 were regarded as dominant subclades with abundances >10%, whereas C163b, C3d and D2 were subdominant with abundances ranging from 1–10%. In Montipora spp., a total of 8 subclades, C116, C15h, C17, C35, C26, C2r, C31 and C3d, could become dominant dependent on the location. Based on an overview of Fig. 3, although Symbiodinium in Montipora spp. did not differ much across regions at the clade level, M. monasteriata exhibited more different Symbiodinium subclade compositions in two sites of Sanya (LHT and SB), whereas Montipora spp. still displayed very similar Symbiodinium subclade compositions in two sites of Hong Kong (CB and LI), supporting the notion that the coral-algal symbiosis is very flexible across host species and geographic distance. The non-parametric multivariate analysis of similarity (ANOSIM) demonstrated that Symbiodinium community structures differed significantly between G. fascicularis and Montipora spp., as well as between coral and seawater samples (R > 0.8, P = 0.01). Moreover, the Symbiodinium community structures among the three regions (Hong Kong, Sanya, and Sansha) were significantly different (R > 0.69, P ≤ 0.03), whereas less significant variations were observed between sampling sites in the same region (CB and LI (Hong Kong), R = 0.257; LHT and SB (Sanya), R = 0.559). These findings indicated a possible environmental adaption across regions and strict host specificity of the Symbiodinium community. Furthermore, most of the subclades identified in corals were also found in the surrounding seawater, suggesting the free-living ability of Symbiodinium and a possible horizontal transmission of Symbiodinium between seawater and corals. Some dominant subclades demonstrated much higher relative abundances in corals compared with those in seawater, suggesting an enrichment of certain Symbiodinium subclades in corals, which is consistent with the findings at the clade level.
Figure 3. Symbiodinium community structures at the subclade level.
Relative abundance of dominant Symbiodinium subclades from G. fascicularis, Montipora spp., and seawater samples in the South China Sea. The color scale on the top represents the percentage of a subclade from each sample. The color scale on the right represents samples from different sites, blue, grey, green, red and purple represent samples from Crescent Bay (CB, Hong Kong), Lamma Island (LI, Hong Kong), Luhuitou (LHT, Sanya), Sunny Bay (SB, Sanya), Drummond Island (DI, Sansha), respectively. Among the selected Symbiodinium subclades, C17.2, C26.b1, and C21 are equivalent to C17, C35, and C3d, respectively. C26a and C35a are both equivalent to C26.
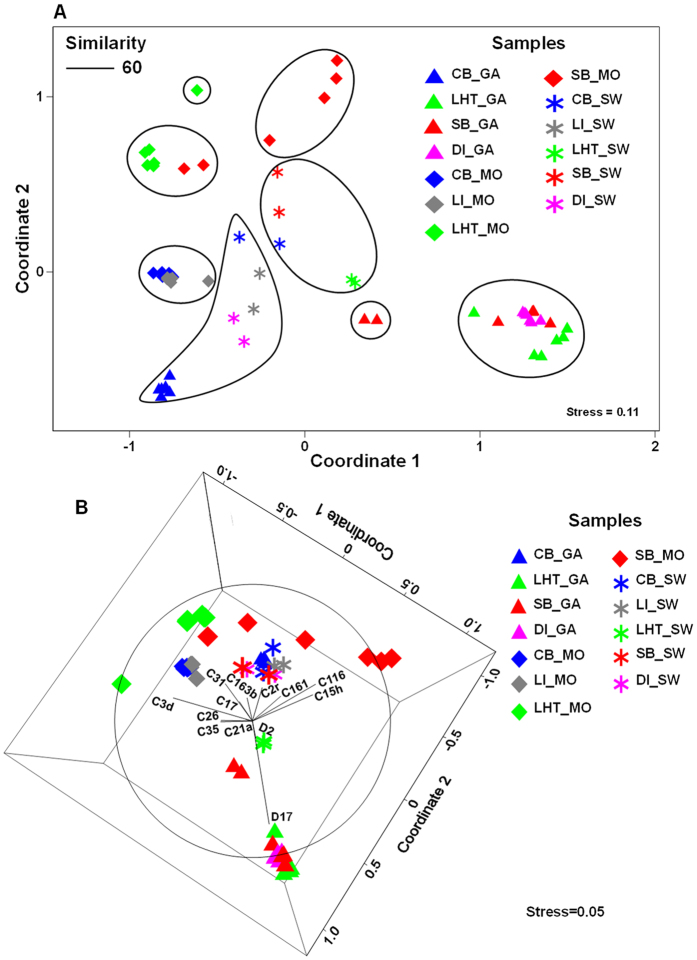
The results of non-metric multidimensional scaling (nMDS) revealed a clear clustering pattern, supporting the dissimilarities among all samples (Fig. 4). All of the seawater samples were located between the G. fascicularis and Montipora spp. samples. Based on a similarity of 60% (Fig. S1, Supporting Information), the Symbiodinium communities of G. fascicularis in Hong Kong were quite different from those in Sanya and Sansha. The Symbiodinium communities of Montipora spp. were less different within the same region (CB and LI (Hong Kong), LHT and SB (Sanya)), but exhibited more differences between different regions (Hong Kong and Sanya). G. fascicularis in Hong Kong and all Montipora spp. were dominated by diverse C subclades, whereas G. fascicularis in Sanya and Sansha mainly harbored two D subclades.
Figure 4. nMDS ordinations.
nMDS plot of G. fascicularis and Montipora spp. dominant Symbiodinium subclade community compositions based on the square root-transformed relative abundance matrix by the Bray-Curtis measure of dissimilarity. Each symbol represents a single sample. Kruskal’s stress numbers are provided. (A): 2dnMDS, with grouping based on complete linkage cluster analysis of 60% similarity. (B): 3dnMDS, with a dominant Symbiodinium subclade overlay vector.
In summary, different coral species at the same sampling site appeared to prefer different Symbiodinium subclades. Congeneric corals from different regions also tended to have significantly distinct Symbiodinium community compositions despite fewer changes in Symbiodinium community compositions of the surrounding seawater. These results suggested that, for congeneric corals, coral-algal symbiosis change mainly resulted from environmental changes across the investigated regions.
Correlations between environmental factors and Symbiodinium communities
According to the canonical correspondence analysis (CCA), temperature, salinity and NH4+ served as the main factors among all potential impact factors (Fig. S2, Supporting Information) affecting Symbiodinium subclades and community compositions, explaining 81.8% and 52.7% of the total subclade variations in G. fascicularis (Fig. 5A) and Montipora spp. (Fig. 5B), respectively. This finding indicated that these three factors provided the largest contribution to coral-algal symbiosis change in the present study. Remarkably, temperature was the most significant factor in shaping subclade compositions, explaining 83.2% and 60.1% of the explanatory variations in G. fascicularis and Montipora spp., respectively. This finding suggested that coral-algal symbiosis change in studied samples was largely driven by temperature. In G. fascicularis, two D subclades (D2, D17) were positively correlated to temperature, while most C subclades had negative correlation with temperature, indicating that G. fascicularis was more likely to select heat-tolerant Symbiodinium with an increasing temperature because the D clade is believed to be more tolerant to heat21,22. In Montipora spp., only three C subclades (C2r, C3d and C163b) were negatively correlated to temperature, whereas the other dominant subclades were all correlated positively to temperature. In both plots, samples from Hong Kong (CB, LI) were clearly separated from those from Sanya and Sansha along axis 1, which was highly correlated to temperature, indicating that temperature is a major factor in shaping Symbiodinium community structures between subtropical (Hong Kong) and tropical (Sanya and Sansha) regions. Thus, temperature provided the greatest environmental contribution to the coral-algal change between subtropical and tropical regions. However, the samples from the tropical region were divided along axis 2, which was mainly correlated to kinds of nutrients, suggesting that nutrients are minor environmental factors affecting Symbiodinium community structures under similar temperature conditions.
Figure 5. Relationships among environmental factors, Symbiodinium subclades, and sampling sites.
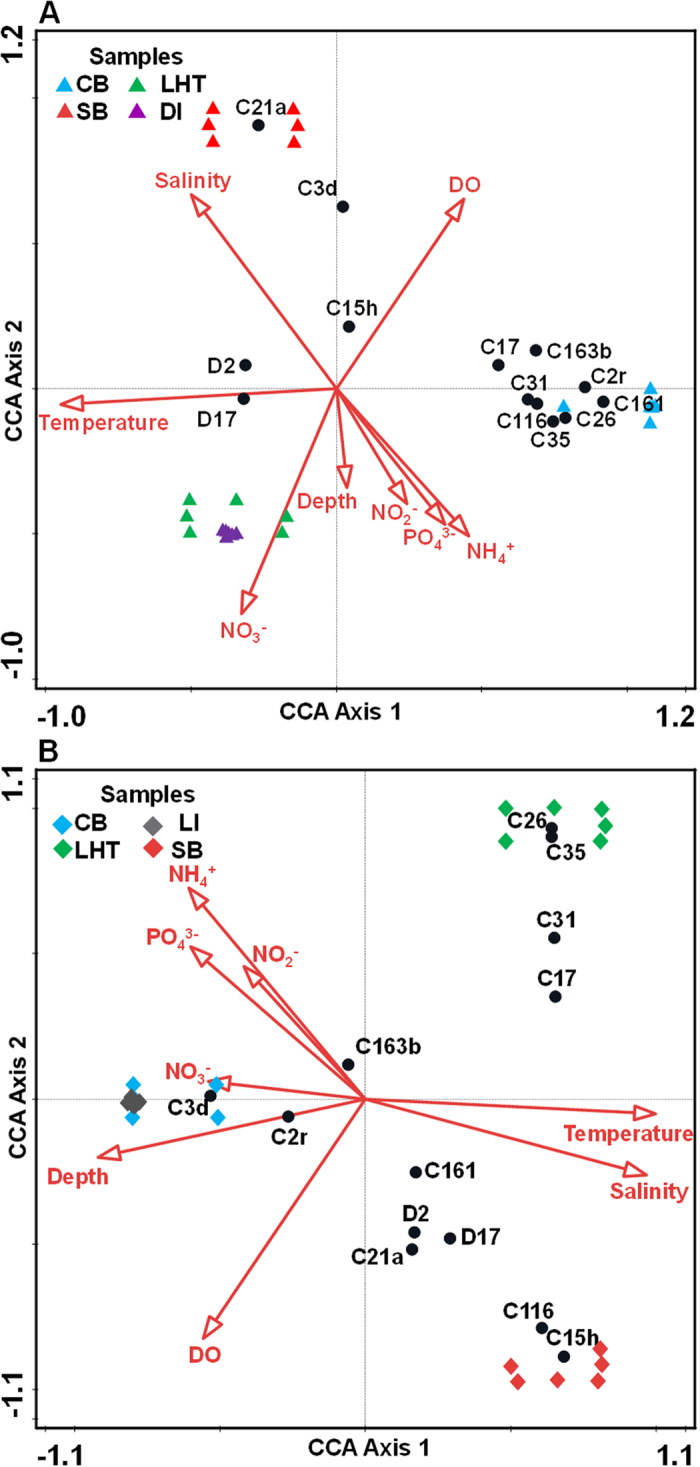
CCA indicates the relationship among environmental factors, Symbiodinium subclades, and coral Symbiodinium community composition from different sampling sites. (A) CCA of G. fascicularis samples. The first axis (CCA Axis 1) explains 75.88% of the total variation and 92.71% of the fitted variation; the second axis (CCA Axis 2) explains 5.96% of the total variation and 7.28% of the fitted variation. (B) CCA of Montipora spp. samples. CCA Axis 1 explains 31.72% of the total variation and 60.18% of the fitted variation; CCA Axis 2 explains 20.89% of the total variation and 39.63% of the fitted variation.
Phylogenetic relationships of dominant Symbiodinium subclades from G. fascicularis and Montipora spp. in the three regions
Symbiodinium subclades dominating congeneric corals tended to be closely distributed in the phylogenetic tree (Fig. 6). For example, C161, C2r, and C163b from G. fascicularis in Hong Kong had closer phylogenetic relationships with D2/D17 from G. fascicularis in Sanya and Sansha compared with the other C subclades that were symbiotic with Montipora spp., suggesting a possible coral-algal coevolution. For the congeneric corals, Symbiodinium associated with corals with a closer geographic distance demonstrated closer phylogenetic relationships, revealing that the phylogenetic relationships of Symbiodinium were linked to geographical distances.
Figure 6. Phylogenetic tree of Symbiodinium subclades that are dominant in G. fascicularis and Montipora spp. in the South China Sea.
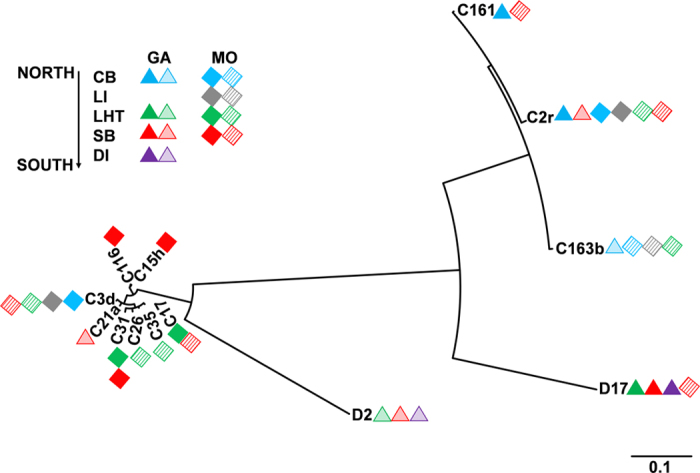
Every symbol represents a group in which the average relative abundance of a certain subclade is >1%. Solid and non-solid symbols indicate average relative abundances of >10% and 1–10%, respectively. The phylograms were analyzed with ITS2 sequences using maximum likelihood and Bayesian inference analyses, and nearly identical profiles were obtained. The presented tree is derived from Bayesian inference.
Discussion
Here, we presented the first systematic investigation of Symbiodinium communities in different coral species across tropical and subtropical regions and indicated the highly spatial flexibility of coral-algal symbiosis in G. fascicularis and Montipora spp. in the South China Sea. Although several previous studies have investigated Symbiodinium community structures of certain scleractinian corals in the South China Sea23,24, this is the first study to demonstrate that, among all potential environmental factors, temperature plays the major role in shaping Symbiodinium community structures and coral-algal symbiosis. The results of the present study are consistent, in part, with some previous findings. Temperature9,25,26, nutrients27, and depth28 all have the potential to affect either the coral host or Symbiodinium community structures, but none of those studies compared the relative effects of these factors or their effects on different coral species. Cooper et al.11 showed, mud content, carbonate content, SST and depth together explained 51.3% of the total variations in dominant Symbiodinium communities in A. millepora and long-term SST explained 10.8% of the total variations. However, they only examined Symbiodinium C1, C2 and D1, whereas we detected 131 Symbiodinium subclades and 13 dominant Symbiodinium subclades in the present study, suggesting a more in-depth coverage of the Symbiodinium communities.
Temperature shapes the structures of the Symbiodinium community
The concordance of the taxonomic data with phylogenetic and ecological evidence led us to propose that, among all potential environmental factors, temperature acted as the major environmental factor impacting coral-algal symbiosis spatial change in the South China Sea. The important role of temperature and the underlying mechanisms can be partly explained by previous empirical evidence. For example, extremely high temperatures can lead to coral bleaching29, but corals that contained more heat-tolerant Symbiodinium were less likely to bleach and bleached corals gained more heat-tolerant Symbiodinium after recovery22,30. Coral larvae gained different Symbiodinium when treated with different temperatures31, whereas adult corals acquired more heat-tolerant Symbiodinium in response to thermal stress9. Additionally, corals from some extremely hot regions have been found to carry some highly heat-tolerant Symbiodinium15. These findings support the conclusion that temperature is a significant factor affecting coral-algal symbiosis. Using a network analysis of stressor interactions, Ban et al.12 suggested that sedimentation, storms and temperature were the most influential stressors in the coral reef ecosystem, which also indicated that temperature could drive coral-algal symbiosis change and play an important role in the establishment of coral-algal symbiosis.
Hong Kong environmental data was collected from HKEPD which was monitored less than one week before the sampling time. As Symbiodinium community in a given coral colony can remain relatively stable in a period of time32, results shall not be affected much. Sansha sampling was finished two months later than the others, it would cause problems when comparing communities between different sites, while Symbiodinium communities in G. fascicularis from Sansha were still highly similar with those in G. fascicularis from Sanya, suggesting Symbiodinium communities in G. fascicularis from Sansha were relatively stable during the two months. The high dissimilarities between Symbiodinium communities from Hong Kong and Sansha/Sanya due to temperature could result from either spatial or temporal variations, as averaged SST in Hong Kong rose ~2 °C/~10 °C from March to April (Sanya sampling)/June (Sansha sampling), respectively. It’s possible that if Hong Kong and Sanya samplings are both finished in June, there would be less differences between Symbiodinium communities in G. fascicularis from Hong Kong and Sansha, and Symbiodinium communities in G. fascicularis from Sanya and Sansha would be more similar. However, spatial variation still played a more important role in the present study: Symbiodinium communities in G. fascicularis from Sanya and Sansha were highly similar even Sansha sampling was two months late; while Symbiodinium communities in G. fascicularis from Hong Kong and Sanya were quite different even sampled at the similar dates. The results of the relationships between Symbiodinium communities and environmental conditions would not be affected by sampling at different time points as long as the environmental data can well reflect the environmental conditions at the sampling points.
Potential roles of Symbiodinium in the coral host in response to bleaching and future climate change
Our results support an argument of coevolution between the coral host and Symbiodinium because Symbiodinium from congeneric corals demonstrated closer phylogenetic relationships and a geographic specificity was observed in the Symbiodinium phylogeny. While we found three C subclades have closer phylogenetic relationships with D subclades than other C subclades, it might result from classification errors, as perfect matches of C2r and C163b were not found in NCBI database and the perfect match of C161 was not identified as Symbiodinium C clade in NCBI. Symbiodinium clade C is believed to possess a higher photosynthetic efficiency, whereas Symbiodinium clade D is more heat tolerant33. Therefore, it can be assumed that the Symbiodinium may contribute to the host adaptation and survival in response to fluctuating environmental conditions. Dixon et al.34 found that Acropora millepora larvae derived from warmer locations possessed greater thermal tolerance, indicating that thermal tolerance was heritable34. Cooper et al.11 observed that adult A. millepora acquired a greater number of heat-tolerant Symbiodinium in response to higher temperatures, suggesting that a potentially thermal-tolerant coral would prefer thermal-tolerant Symbiodinium subclades during growth to facilitate their future survival during exposure to relatively high temperatures. However, Palumbi et al.35 found that A. hyacinthus became more heat tolerant without changes in symbiont clade composition as well as gene expression35. Thus, different coral species may possess different heat tolerance mechanisms.
Tchernov et al.36 showed that a rise in temperature damaged the membranes of free-living Symbiodinium and thus disrupted their photosynthetic ability, ultimately leading to death, and that the compositions of membranes determined the thermal stress sensitivity of Symbiodinium36. In the present study, we found that as the temperature rose across the regions, the relative abundance of some heat-sensitive Symbiodinium C subclades, such as C116, C17, decreased in seawater. In contrast, some Symbiodinium that are supposed to be heat sensitive according to the clade characteristics, such as C31, were not greatly affected much across the regions and C31 was found to have a high relative abundance in Montipora spp. from Sanya. These findings indicated that some Symbiodinium might possess compensation mechanisms to survive under thermal stress that could be taken advantage by the coral host to adapt to future climate change. However, previous studies have shown that some coral species acquired their Symbiodinium, e.g., C31, through vertical transmission37,38. In the present study, we also detected C15h, C21a, C35 and C26 in coral samples but not in ambient seawater samples in certain locations, either due to a potential vertical transmission or to an insufficient sensitivity of the current techniques. We suspect that if a vertically transmitted Symbiodinium fails to develop compensation mechanisms, coral host would become more susceptible to future climate change.
G. fascicularis was found to have greater heat tolerance than Montipora sp.39, and similarly in our study, very few members of the Symbiodinium D clade were detected among the Montipora spp., even in Sanya, where temperature is relatively high. This observation raises the possibility that heat-tolerant Symbiodinium facilitates coral adaptation. In response to global warming with a predicted increase in bleaching events, coral species that can acquire more heat-tolerant Symbiodinium will survive better. Coral species that lack the symbiotic flexibility with Symbiodinium subclades have also been found to be more susceptible to bleaching29. However, some heat-tolerant Symbiodinium can reduce host calcification and photosynthetic efficiency, which may impair the competitive ability of the coral26,33. While Cunning et al.40 demonstrated that the growth disadvantage of heat-tolerant Symbiodinium could be reduced by elevated temperature, thus enhancing the survival of coral host. More investigations are needed to decipher the mechanisms that how coral species with different Symbiodinium survive during future climate change.
Pettay et al.33 found that Symbiodinium trenchii invaded the Caribbean and noted that this Symbiodinium might drive coral-algal symbiosis dynamics during climate change, which is inconsistent with our findings. According to the present study, all Symbiodinium (including Symbiodinium trenchii) could potentially be transmitted between corals and surrounding seawater, and certain subclades of Symbiodinium were enriched in the coral host. For certain Symbiodinium subclades such as C161, congeneric corals in different regions surrounded by a similar relative abundance of certain Symbiodinium are internally enriched with different relative abundances of that Symbiodinium. Therefore, the acquisition of Symbiodinium by corals shall not be a random but a selective event. Likely, corals are capable of selecting certain Symbiodinium that are potentially more beneficial to them, and the selected Symbiodinium facilitate the adaptation of the host. Thus, the coral host drives the dynamics of the symbiosis. However, we cannot exclude the possibility that, in certain coral species, invading Symbiodinium may drive the dynamics of the symbiosis.
Ecological implications
The adaptation of the coral reef ecosystem to future climate change strongly relies on the adaptation of their internal Symbiodinium community. In the South China Sea, among all potential environmental factors, temperature can serve as a major factor for shaping coral-algal symbiosis. Therefore, a rise in SST is a prominent threat to the South China Sea corals, but still corals have the potential to adapt to future climate change with their flexibility of symbiosis with Symbiodinium. Although more caution should be adhered to when transplanting a new species into a region33, the introduction of more heat-tolerant Symbiodinium into the coral reef ecosystem may facilitate their adaptation to the rising SST41.
Previous studies have shown that heat-tolerant corals contain a greater number of clade D Symbiodinium42. Although both G. fascicularis and Montipora spp. have the potential to change their Symbiodinium community structures in response to environmental changes, G. fascicularis appears to prefer more heat-tolerant Symbiodinium subclades, which may lead to its dominance during future climate change. Indeed, in recent years some dominant coral species have been declining while some subdominant coral species are becoming more dominant29,43. During the development of temperature-driven symbiosis, the flexibility of coral-algal symbiosis and preference for certain Symbiodinium of coral species can directly determine coral species’ competition during future climate change, which will ultimately affect future coral population structures.
However, human activities such as seawater pollution, overfishing, and tourism etc. have also caused a rapid decline in the coral cover44,45. In the present study, nutrients had only a marginal impact on coral-algal symbiosis, considering nutrients contribute the most to coral cover and species richness loss compared with other water quality parameters46, suggesting a lack of symbiotic flexibility in response to seawater pollution such as eutrophication. Increases in coastal nutrients are frequently linked to human activities47,48, which suggests that human activities can also affect coral-algal symbiosis. This observation indicates that seawater pollution may be another important threat to the coral reef.
Conclusions
The present study demonstrated that temperature drove coral-algal symbiosis spatial change in the South China Sea and corals had the potential to adapt to future climate change with selecting more heat-tolerant Symbiodinium under gradually rising SST. Thus, for the future conservation of corals in the South China Sea, the introduction to coral hosts of heat-tolerant Symbiodinium might facilitate their adaptation to future climate change. Despite the high level of spatial flexibility of coral-algal symbiosis in G. fascicularis and Montipora spp. in the South China Sea, future studies are needed on a larger geographic scale and shall be expanded to other dominant coral species like Acropora spp. to further address coral-algal symbiosis in response to future climate change.
Methods
Sample collection and preparation
Forty-eight coral samples of G. fascicularis (n = 24 colonies) and Montipora spp. (n = 24 colonies) and 10 seawater samples in total were collected from two Hong Kong sampling sites in March 2014, two Sanya sampling sites in April 2014 and one Sansha sampling site in June 2014 (Table 1). At each sampling site, one small piece (~1 cm × 1 cm) from each healthy coral colony was collected using a hammer/chisel set and wrapped in a tagged bag filled with seawater. On board, the coral pieces were immediately washed with filtered seawater, fixed in 70% ethanol, stored and transported to the laboratory using a cool box containing dry ice. The seawater surrounding the coral colonies was also collected as controls. Free-living Symbiodinium cells in seawater were collected by filtering seawater through 0.22-μm polycarbonate membranes and fixed with 50% ethanol. All of the fixed coral and seawater samples were stored at −30 °C until DNA extraction.
Environmental data collection
Hong Kong environmental data were provided by the Hong Kong Environmental Protection Department (HKEPD). Data for the sampling month from five selected monitoring stations around each sampling site (Table S1, Table S2, Supporting Information) were collected and averaged, and the averages were used as environmental factor data for a given sampling site. For Sanya and Sansha, the seawater temperature, salinity and depth were measured during the sampling in situ with CTD (Idronaut, Italy), while dissolved oxygen (DO) was assessed in situ using a YSI 6600V2-02 multi-parameter instrument (YSI, USA). The seawater samples were collected and immediately filtered (Whatman GF/F, 47 mm) to analyze dissolved nutrients (nitrite, nitrate, ammonia and phosphate) using a Lachat QC8500 Flow Injection Autoanalyzer (Lachat Instruments, USA).
DNA extraction and amplicon sequencing
A small fragment (~0.5 cm × 0.5 cm) cut from each fixed piece of coral was first rinsed with 1*PBSE (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4 and 10 mM EDTA) and then mashed in 1*PBSE using a mortar and a pestle. The mashed coral serous was centrifuged at 12000 g, and the pellets were collected for DNA extraction. DNA was extracted from each pellet using the FastDNA® Spin Kit for Soil (MP Biomedicals, France) following the protocol provided with the kit. After quality and purity examinations, the extracted DNA samples were applied as PCR templates. The Symbiodinium ITS2 region of the rDNA was amplified by PCR using primers (F: 5′GAATTGCAGAACTCCGTG-3′; R: 5′ GGATCCATATGCTTAAGTTCAGCGGGT-3′) designed to produce 330–360 bp ITS2 fragments49. At the 5′ terminus of the forward primer, six-nucleotide unique barcodes were attached for multiplexed sequencing. The PCR amplifications were conducted in a 50-μL reaction volume containing ~50 ng of DNA, 25 μL of 2× Taq Platinum PCR Master (Tiangen, China), 200 nM of each primer, and ddH2O up to the final volume. The reactions were performed under the following conditions: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 51 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. Each sample was amplified in three independent reactions to minimize potential PCR bias, and then purified using the PureLink® PCR Purification Kit (Invitrogen, USA). The purified PCR products were quantified using a Thermo NanoDrop 2000 UV-Vis Spectrophotometer and then mixed based on equal mass for subsequent multiplexed amplicon sequencing. The final DNA samples were sequenced following a paired-end (PE) 300 bp × 2 strategy on an Illumina MiSeq sequencer operated by the Novogene company (Beijing, China). The sequencing datasets were submitted to the NCBI Sequence Read Archive under accession number SRP066283.
Data processing, ITS2 database establishment and data analysis
Strict quality control and sequence filtration were applied to ensure the accuracy of the following analysis. Adaptors, short reads, and low quality reads were first removed by the sequencing company. The PEAR (paired-end read merger) tool was applied to obtain full-length ITS2 rDNA fragments with merging overlapping PE reads to generate ITS2 sequences50 (http://sco.h-its.org/exelixis/web/software/pear/, see commands in Supporting Information). ITS2 tags were demultiplexed into all samples in the QIIME platform51 by identifying unique barcodes (http://qiime.org/index.html, see commands in Supporting Information). We found that the previous ITS2 database included some duplicate sequences52, therefore we uploaded the previous ITS2 database to CD-HIT Suite website (http://weizhongli-lab.org/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit-est), set sequence identity cut-off as 100%, compared both strands, set all other parameters as default53, removed the duplicates, merged the annotations and used the results to establish a non-redundant ITS2 database (ITS2 Database.doc, Supporting Information). Next, all of the sequences were aligned to the ITS2 database by BLASTN54 based on E-value of 1e-5 (see commands in Supporting Information), the output were then filtered in Microsoft Excel (Microsoft Corp., USA) based on 97% similarity for Symbiodinium subclade identification. For ITS2 rDNA, 97% sequence similarity has been proved to be reliable for Symbiodinium subclade classification and could permit differences of ten base pairs for identification within the same Symbiodinium subclade as a consequence of intra-genomic sequence divergence10.
Thirteen (sub)dominated Symbiodinium subclades were picked from 131 total aligned Symbiodinium subclades for following analysis. The relative abundances of picked Symbiodinium subclades were >10% in at least one sample or their average relative abundances in all samples were at least around 1% and the filtering results can indicate the original communities well (whole_community_data.xls, Supporting Information). To profile Symbiodinium communities in different samples, a heat map and scatter plots were created with OriginPro 2015 (OriginLab, USA). To justify significances between Symbiodinium community structures across the sampling sites, a two-way ANOSIM and a permutational multivariate analysis of variance (PERMANOVA) were performed using PRIMER 7 software55. To present the relationships of the Symbiodinium community structures from different sampling sites, a nMDS ordination as well as clustering analysis were developed based on the square root-transformed Bray-Curtis similarity matrix of Symbiodinium profiles (Symbiodinium relative abundance) in PRIMER. To test the relationships among environmental factors, Symbiodinium community, and sampling sites, CCA was conducted in Canoco 556. CCA uses ecological data sets to extract synthetic environmental gradients which are the basis for revealing the differential habitat preferences of microbes and has been proved to be reliable when combining spatial and seasonal sampling together, as long as the sample size is granted and the environment conditions can reflect habitat preference of the communities57,58. To identify the phylogenetic relationships among the dominant Symbiodinium subclades found in this study, two phylogenetic trees were constructed based on the Kimura 2-parameter model with uniform rates among sites using Maximum Likelihood in MEGA 659 and Bayesian Inference in Mrbayes60, respectively.
Additional Information
How to cite this article: Tong, H. et al. Temperature shapes coral-algal symbiosis in the South China Sea. Sci. Rep. 7, 40118; doi: 10.1038/srep40118 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the grant of NSFC-Guangdong Joint Fund (# U1301232) to PY Qian. The authors thank Dr. Xiubao Li and Dr. Sheng Liu for many helpful discussions.
Footnotes
Author Contributions H.T. and P.Y.Q. wrote the paper. H.T., L.C., G.Z., T.Y., R.T. and W.Z. performed the research. H.T. analyzed the data. P.Y.Q and H.H. designed the research. All authors reviewed the paper.
References
- Carpenter K. E. et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563, doi: 10.1126/science.1159196 (2008). [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. et al. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742, doi: 10.1126/science.1152509 (2007). [DOI] [PubMed] [Google Scholar]
- Muscatine L., Falkowski P. G., Porter J. W. & Dubinsky Z. Fate of photosynthetic fixed carbon in light- and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc R Soc Ser B-Bio 222, 181–202, doi: 10.1098/rspb.1984.0058 (1984). [DOI] [Google Scholar]
- Pochon X. & Gates R. D. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol Phylogenet Evol 56, 492–497, doi: 10.1016/j.ympev.2010.03.040 (2010). [DOI] [PubMed] [Google Scholar]
- Baker A. C. Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol S 34, 661–689, doi: 10.1146/annurev.ecolsys.34.011802.132417 (2003). [DOI] [Google Scholar]
- Silverstein R. N., Correa A. M. S. & Baker A. C. Specificity is rarely absolute in coral-algal symbiosis: implications for coral response to climate change. Proc R Soc Ser B-Bio 279, 2609–2618, doi: 10.1098/rspb.2012.0055 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog J. C., van Oppen M. J. H., Cantin N. E., Stam W. T. & Olsen J. L. Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26, 449–457, doi: 10.1007/s00338-007-0244-8 (2007). [DOI] [Google Scholar]
- Ziegler M., Roder C. M., Buchel C. & Voolstra C. R. Limits to physiological plasticity of the coral Pocillopora verrucosa from the central Red Sea. Coral Reefs 33, 1115–1129, doi: 10.1007/s00338-014-1192-8 (2014). [DOI] [Google Scholar]
- Silverstein R. N., Cunning R. & Baker A. C. Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Global Change Biology 21, 236–249, doi: 10.1111/gcb.12706 (2015). [DOI] [PubMed] [Google Scholar]
- Thomas L., Kendrick G. A., Kennington W. J., Richards Z. T. & Stat M. Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Molecular Ecology 23, 3113–3126, doi: 10.1111/mec.12801 (2014). [DOI] [PubMed] [Google Scholar]
- Cooper T. F. et al. Environmental factors controlling the distribution of symbiodinium harboured by the coral Acropora millepora on the Great Barrier Reef. PLoS One 6, e25536, doi: 10.1371/journal.pone.0025536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban S. S., Graham N. A. & Connolly S. R. Evidence for multiple stressor interactions and effects on coral reefs. Global Change Biology 20, 681–697, doi: 10.1111/gcb.12453 (2014). [DOI] [PubMed] [Google Scholar]
- Hoeghguldberg O. & Smith G. J. Influence of the population density of zooxanthellae and supply of ammonium on the biomass and metabolic characteristics of the reef corals Seriatopora hystrix and Stylophora pistillata. Mar Ecol Prog Ser 57, 173–186, doi: 10.3354/meps057173 (1989). [DOI] [Google Scholar]
- Klepac C. N. et al. Seasonal stability of coral-Symbiodinium associations in the subtropical coral habitat of St. Lucie Reef, Florida. Mar Ecol Prog Ser 532, 137–151, doi: 10.3354/meps11369 (2015). [DOI] [Google Scholar]
- D’Angelo C. et al. Local adaptation constrains the distribution potential of heat-tolerant Symbiodinium from the Persian/Arabian Gulf. International Society for Microbial Ecology, doi: 10.1038/ismej.2015.80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodkin N. F. et al. Coral communities of Hong Kong: long-lived corals in a marginal reef environment. Mar Ecol Prog Ser 426, 185–196, doi: 10.3354/meps09019 (2011). [DOI] [Google Scholar]
- Yeung C. W. et al. Environmental variabilities and the distribution of octocorals and black corals in Hong Kong. Marine Pollution Bulletin 85, 774–782, doi: 10.1016/j.marpolbul.2013.12.043 (2014). [DOI] [PubMed] [Google Scholar]
- Qiu W. F. The Sanya Coral Reef National Marine Nature Reserve, China: A governance analysis. Mar Policy 41, 50–56, doi: 10.1016/j.marpol.2012.12.030 (2013). [DOI] [Google Scholar]
- Li X. B. et al. Spatial and temporal variations in sediment accumulation and their impacts on coral communities in the Sanya Coral Reef Reserve, Hainan, China. Deep-Sea Res Pt Ii 96, 88–96, doi: 10.1016/j.dsr2.2013.04.015 (2013). [DOI] [Google Scholar]
- Huang H. et al. Coral cover as a proxy of disturbance: A case study of the biodiversity of the hermatypic corals in Yongxing Island, Xisha Islands in the South China Sea. Chinese Sci Bull 51, 129–135, doi: 10.1007/s11434-006-9129-4 (2006). [DOI] [Google Scholar]
- LaJeunesse T. C., Smith R. T., Finney J. & Oxenford H. Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proc R Soc Ser B-Bio 276, 4139–4148, doi: 10.1098/rspb.2009.1405 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. W., Hernandez-Pech X., Iglesias-Prieto R., Fitt W. K. & Schmidt G. W. Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol Oceanogr 59, 788–797, doi: 10.4319/lo.2014.59.3.0788 (2014). [DOI] [Google Scholar]
- Zhou G., Huang H., Lian J., Zhang C. & Li X. Habitat correlation of Symbiodinium diversity in two reef-building coral species in an upwelling region, eastern Hainan Island, China. Journal of the Marine Biological Association of the United Kingdom 92, 1309–1316, doi: 10.1017/s0025315411001548 (2011). [DOI] [Google Scholar]
- Huang H. et al. Latitudinal variation in algal symbionts within the scleractinian coral Galaxea fascicularis in the South China Sea. Marine Biology Research 7, 208–211, doi: 10.1080/17451000.2010.489616 (2011). [DOI] [Google Scholar]
- Rodolfo-Metalpa R. et al. Thermally tolerant corals have limited capacity to acclimatize to future warming. Global Change Biology 20, 3036–3049, doi: 10.1111/gcb.12571 (2014). [DOI] [PubMed] [Google Scholar]
- Cunning R., Silverstein R. N. & Baker A. C. Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc R Soc Ser B-Bio 282, 20141725, doi: 10.1098/rspb.2014.1725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawall Y., Al-Sofyani A., Banguera-Hinestroza E. & Voolstra C. R. Spatio-Temporal Analyses of Symbiodinium Physiology of the Coral Pocillopora verrucosa along Large-Scale Nutrient and Temperature Gradients in the Red Sea. PLoS One 9, doi: 10.1371/journal.pone.0103179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. S. The engine of the reef: photobiology of the coral-algal symbiosis. Frontiers in Microbiology 5, 422, doi: 10.3389/fmicb.2014.00422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grottoli A. G. et al. The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Global Change Biology 20, 3823–3833, doi: 10.1111/gcb.12658 (2014). [DOI] [PubMed] [Google Scholar]
- LaJeunesse T. C. et al. Host-symbiont recombination versus natural selection in the response of coral-dinoflagellate symbioses to environmental disturbance. Proc R Soc Ser B-Bio 277, 2925–2934, doi: 10.1098/rspb.2010.0385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler N. S., Pandolfi J. M. & Sampayo E. M. Symbiodinium identity alters the temperature-dependent settlement behaviour of Acropora millepora coral larvae before the onset of symbiosis. Proc R Soc Ser B-Bio 282, 20142260, doi: 10.1098/rspb.2014.2260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinley M. P. et al. Symbiodinium spp. in colonies of eastern Pacific Pocillopora spp. are highly stable despite the prevalence of low-abundance background populations. Mar Ecol Prog Ser 462, 1–7 (2012). [Google Scholar]
- Pettay D. T., Wham D. C., Smith R. T., Iglesias-Prieto R. & LaJeunesse T. C. Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proceedings of the National Academy of Sciences of the United States of America 112, 7513–7518, doi: 10.1073/pnas.1502283112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon G. B. et al. Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462, doi: 10.1126/science.1261224 (2015). [DOI] [PubMed] [Google Scholar]
- Palumbi S. R., Barshis D. J., Traylor-Knowles N. & Bay R. A. Mechanisms of reef coral resistance to future climate change. Science 344, 895–898, doi: 10.1126/science.1251336 (2014). [DOI] [PubMed] [Google Scholar]
- Tchernov D. et al. Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proceedings of the National Academy of Sciences of the United States of America 101, 13531–13535, doi: 10.1073/pnas.0402907101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse T. C. et al. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596–603, doi: 10.1007/s00338-004-0428-4 (2004). [DOI] [Google Scholar]
- Padilla-Gamino J. L., Pochon X., Bird C., Concepcion G. T. & Gates R. D. From Parent to Gamete: Vertical Transmission of Symbiodinium (Dinophyceae) ITS2 Sequence Assemblages in the Reef Building Coral Montipora capitata. PLoS One 7, doi: 10.1371/journal.pone.0038440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P. A. & Baird A. H. Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19, 155–163, doi: 10.1007/s003380000086 (2000). [DOI] [Google Scholar]
- Cunning R., Gillette P., Capo T., Galvez K. & Baker A. C. Growth tradeoffs associated with thermotolerant symbionts in the coral Pocillopora damicornis are lost in warmer oceans. Coral Reefs 34, 155–160 (2015). [Google Scholar]
- van Oppen M. J., Oliver J. K., Putnam H. M. & Gates R. D. Building coral reef resilience through assisted evolution. Proceedings of the National Academy of Sciences of the United States of America 112, 2307–2313, doi: 10.1073/pnas.1422301112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis D. J. et al. Genomic basis for coral resilience to climate change. Proceedings of the National Academy of Sciences of the United States of America 110, 1387–1392, doi: 10.1073/pnas.1210224110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. E., Dunne R. P., Phongsuwan N., Patchim L. & Hawkridge J. M. The reef coral Goniastrea aspera: a ‘winner’ becomes a ‘loser’ during a severe bleaching event in Thailand. Coral Reefs 33, 395–401, doi: 10.1007/s00338-013-1120-3 (2014). [DOI] [Google Scholar]
- Spalding M. D. & Brown B. E. Warm-water coral reefs and climate change. Science 350, 769–771, doi: 10.1126/science.aad0349 (2015). [DOI] [PubMed] [Google Scholar]
- Bellwood D. R., Hoey A. S. & Hughes T. P. Human activity selectively impacts the ecosystem roles of parrotfishes on coral reefs. Proc R Soc Ser B-Bio 279, 1621–1629, doi: 10.1098/rspb.2011.1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprey N. N., Yasuhara M. & Baker D. M. Reefs of tomorrow: eutrophication reduces coral biodiversity in an urbanized seascape. Glob Chang Biol, doi: 10.1111/gcb.13432 (2016). [DOI] [PubMed] [Google Scholar]
- Smith V. H., Tilman G. D. & Nekola J. C. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ Pollut 100, 179–196, doi: 10.1016/S0269-7491(99)00091-3 (1999). [DOI] [PubMed] [Google Scholar]
- Doney S. C. The Growing Human Footprint on Coastal and Open-Ocean Biogeochemistry. Science 328, 1512–1516, doi: 10.1126/science.1185198 (2010). [DOI] [PubMed] [Google Scholar]
- Lajeunesse T. C. & Trench R. K. Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). The Biological Bulletin 199, 126–134 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang J. J., Kobert K., Flouri T. & Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620, doi: 10.1093/bioinformatics/btt593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, doi: 10.1038/nmeth.f.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif C. et al. Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Molecular Ecology 23, 4418–4433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Niu B. F., Gao Y., Fu L. M. & Li W. Z. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682, doi: 10.1093/bioinformatics/btq003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic Local Alignment Search Tool. J Mol Biol 215, 403–410, doi: 10.1006/jmbi.1990.9999 (1990). [DOI] [PubMed] [Google Scholar]
- Clarke K. R. & Gorley R. N. PRIMER v7: User Manual/Tutorial. (PRIMER-E: Plymouth, 2015).
- Šmilauer P. & Lepš J. Multivariate analysis of ecological data using CANOCO 5. (Cambridge university press, 2014). [Google Scholar]
- ter Braak C. J. F. & Verdonschot P. F. M. Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquatic Sciences 57, 255–289, doi: 10.1007/bf00877430 (1995). [DOI] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62, 142–160, doi: 10.1111/j.1574-6941.2007.00375.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol 61, 539–542, doi: 10.1093/sysbio/sys029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.