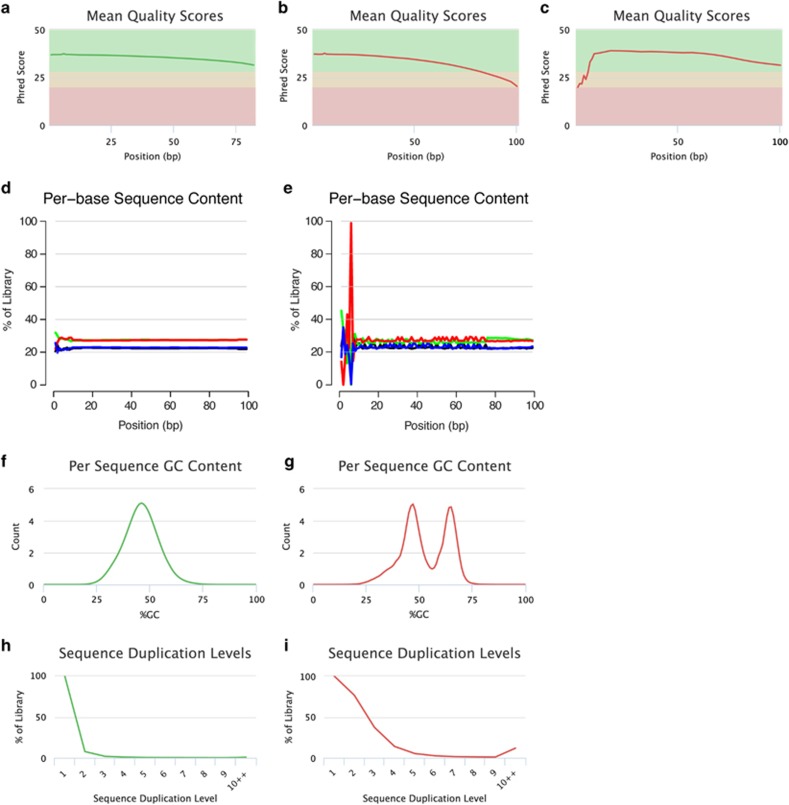
Figure 2.
Read quality assessment. Per-base sequence quality plots indicate the mean quality scores for each nucleotide position in all reads. Background colors highlight the quality of the call (that is, green: high quality; yellow: reasonable quality; red: low quality). Examples: (a) Reads exhibit high base quality scores at each position. (b) Sequenced nucleotides initially exhibit high-quality scores but the per-base read quality decreases with increasing read length, reaching lower quality values toward the read end, necessitating read trimming. (c) Initial low per-base quality that recovers to a high quality later in the run. Under this circumstance, error correction can be applied but trimming is generally not advisable. Per-base sequence content plots indicate the proportion of each nucleotide for each read position. Examples: (d) A random library with little difference of base composition (colors indicate different nucleotides: green: A; blue: C; black: G; red: T) between single read positions. (e) Imbalance of different bases, potentially caused by overrepresented sequences (for example, adapters). Per sequence GC-content plots indicate the observed GC-content of all reads. Examples: (f) The GC-content of the reads is normally distributed with a peak that corresponds to the overall genomic GC-content of the studied species. (g) The bimodal shape of the distribution of the reads' GC-content suggests that the sequenced library may have been contaminated or that adapter sequences may still be present. Sequence duplication plots indicate the level of duplication among all reads in the library (reads with more than 10 duplicates are binned). Examples: (h) The low level of sequence duplication suggests that a diverse library has been sequenced with a high coverage of target sequence. (i) A high level of sequence duplications often suggests either a technical artifact (for example, because of PCR overamplification) or biological duplications. All examples were generated using FastQC and plotted using MultiQC (Ewels et al., 2016).