Abstract
Objective
Previous studies have reported that Ras-associated domain family 1A (RASSF1A), the most commonly silenced tumor suppressor via promoter methylation, played vital roles in the development of carcinogenesis. The purpose of this meta-analysis was to determine whether RASSF1A promoter methylation increased the risk of thyroid cancer.
Methods
PubMed, Embase, ISI Web of Knowledge, and Chinese National Knowledge Infrastructure databases were searched to obtain eligible studies. The pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to estimate the strength of the associations, using Stata 12.0 software. The methodological quality of included studies was evaluated using Newcastle–Ottawa scale table. Egger’s test and Begg’s test were applied to detect publication biases. TSA 0.9 software was used to calculate the required information size and whether the result was conclusive.
Results
A total of 10 articles with 12 studies that included 422 thyroid cancer patients, identifying the association of RASSF1A promoter methylation with thyroid cancer risk, were collected in this meta-analysis. Overall, RASSF1A promoter methylation significantly increased the risk of thyroid cancer (total, OR=8.27, CI=4.38–15.62, P<0.05; Caucasian, OR=9.25, CI=3.97–21.56, P<0.05; Asian, OR=7.01, CI=2.68–18.38, P<0.05). In the subgroup analysis based on sample type, a significant association between thyroid cancer group and control group was found (normal tissue, OR=9.55, CI=4.21–21.67, P<0.05; adjacent tissue, OR=6.80, CI=2.49–18.56, P<0.05). The frequency of RASSF1A promoter methylation in follicular thyroid carcinoma was higher than in control group (OR=11.88, CI=5.80–24.32, P<0.05). In addition, the results indicated that the RASSF1A promoter methylation was correlated with papillary thyroid carcinoma in Caucasians and Asians (total, OR=8.07, CI=3.54–18.41, P<0.05; Caucasian, OR=11.35, CI=2.39–53.98, P<0.05; Asian, OR=6.67, CI=2.53–17.64, P<0.05). On the basis of the trial sequential analysis, the significant association of RASSF1A promoter methylation with thyroid cancer risk was found, and there was no need to perform further studies.
Conclusion
This meta-analysis confirms that RASSF1A promoter methylation is a risk factor for thyroid tumor.
Keywords: RASSF1A, methylation, thyroid neoplasms, meta-analysis
Introduction
It has been reported that thyroid carcinomas, the most frequently reported endocrine neoplasia, account for only 3%–4% in all human tumors, but the incidence of thyroid cancer is steadily increasing and has the highest increase in incidence within the past two decades.1 Currently, the use of neck ultrasonography is widely used in the diagnosis of thyroid cancer and brings light to the detection of many small early-stage tumors.2 However, the incidence of detection of large tumors and advanced stage tumor patients have also increased in these years.3,4 Thyroid cancer can be categorized into four histotypes: follicular thyroid carcinoma (FTC), papillary thyroid carcinoma (PTC), anaplastic thyroid carcinoma, and medullary thyroid carcinoma (MTC). PTC, the most common type of thyroid cancer, represents 80% of patients, followed by FTC (10%).5,6 PTC derives from the follicular cells of the thyroid, and it has a distinctive papillary architecture. In addition, the nuclear membrane of cells in PTC has several distinctive alternations, such as grooves, pseudoinclusions, and optical clearing. In contrast to the papillary carcinomas, in which the change of nuclear membrane alternations is vital, the diagnosis of FTC depends on whether the thyroid tumor has invaded through the surrounding vessels. MTC has a rare incidence accounting for ~3%, which derives from parafollicular C cells.7 Compared with FTC and PTC – well differentiated cancer – MTC has a more aggressive clinical characteristic.8 MTC often occurs in a form of inherited cancer, which is known as multiple endocrine neoplasia type 2.9 In recent decades, although many genetic studies for thyroid cancer have been performed, no specific biomarkers for the large number of sporadic thyroid cancers have been found.10–12
The Ras-associated domain family 1 (RASSF1) family of proteins, representing one type of Ras effectors, can inhibit the development of cancer. RASSF1A, one of the seven iso-forms of the RASSF1 family and located on 3p21, is the most common epigenetically inactivated tumor suppressor genes via promoter methylation in human cancers.13,14 It can bind Ras protein in a guanosine triphosphate-dependent manner to mediate the cell apoptotic, which has a similar function with Nore1 in mouse.14 In many solid tumors, RASSF1A, as a component of crucial cell signal pathways, plays an important role in the Ras/PI3K/AKT, Ras/RAF/MEK/ERK, and Hippo pathways, and the inactivation of RASSF1A can result in the alternations of clinical characteristic in tumors.15,16 It has been reported that the CpG methylation of RASSF1A promoter can lead to the loss-of-expression of RASSF1A.14 Previous studies have found that the RASSF1A promoter methylation can increase the risk of lung cancer, breast cancer, prostate cancer, ovarian cancer, renal cell carcinoma, colorectal cancer, hepatocellular carcinoma, and gastric cancer.17–24 Furthermore, these results indicated that the RASSF1A promoter methylation may be a significant prognostic factor for many human cancers. Despite a number of studies performed on thyroid cancer patients, the relationship of RASSF1A promoter methylation with thyroid cancer risk and pathogenesis remains controversial. To clarify these controversial results, a meta-analysis was performed.
Methods
Publication search
Relevant literature was retrieved from PubMed, Embase, ISI Web of Knowledge, and Chinese National Knowledge Infrastructure databases up until September 2016, without language restrictions. The following keywords were applied: “RASSF1 protein, human (Supplementary Concept),” “Thyroid Neoplasms (Mesh),” “Methylation (Mesh),” “RASSF1A,” “thyroid cancer,” and “hypermethylation” to search eligible studies. Review articles and the references from relevant primary studies were manually searched for identifying additional potential studies.
Inclusion and exclusion criteria
The inclusion criteria were as follows: 1) studies primarily evaluating the frequency of RASSF1A promoter methylation in thyroid cancer with control group, 2) studies that have a case–control design, 3) studies that had sufficient data of frequency, and 4) patients who had been accurately diagnosed according to the diagnostic criteria. Studies were removed if they met one of the items: 1) reviews, meta-analysis, or animal studies; 2) duplicate data; and 3) study performed in the cell lines.
Data extraction
The following data were extracted from the eligible studies: year of publication, country, ethnicity, frequency of RASSF1A promoter methylation, cancer type, method of methylation detection, and sample type. The information was reviewed by two investigators. If the two investigators had divergence, the disagreement was resolved by consulting with the other authors.
Quality assessment
To ensure the high quality of included literatures, the Newcastle–Ottawa scale (NOS) table was applied to assess the methodological quality of observational studies. The NOS is commonly used in the quality assessment of nonrandomized studies, such as case–control and cohort studies. The scores of quality assessment range from 0 to 9. If the study got a score <5 it was excluded.
Trial sequential analysis
In a cumulative meta-analysis, false-positive and false-negative results may arise, which result from the repeated testing and the adding of other studies. Therefore, it was crucial to lower the risk of type I error and type II error in the repeated statistics. In addition, interim analysis was performed to estimate the required sample size according to the data in the common trials and to avoid wasting of the sample. In this present meta-analysis, the trial sequential analysis was performed to estimate the required information size (RIS) and observe whether the result was conclusive through controlling the risk of type I error and type II error. TSA 0.9 software (www.ctu.dk/tsa) was applied to conduct these analyses by combining the RIS with trial sequential monitoring boundaries. The trial sequential analysis was conducted at the level of 5% risk of type I error and 80% power of statistical test (5% risk of type II error). Moreover, adjusted information size was calculated with a relative risk reduction (−50%) according to the incidence of control and case groups.
Statistical analysis
The frequency of RASSF1A promoter methylation in case and control groups was collected to calculate the pooled odds ratios (ORs) and 95% confidence interval (CI),25 evaluating the strength of association of RASSF1A promoter methylation with thyroid cancer risk or thyroid cancer clinical characteristics. Heterogeneity among studies was measured using Cochran’s Q-test and Higgin’s I2.26,27 If heterogeneity existed in studies (P<0.05, I2>50%), random-effects model was applied, otherwise, fixed-effects model was used (P>0.05, I2<50%).28,29 In addition, subgroup analysis based on ethnicity and sample type were conducted to detect the source of heterogeneity and lower the between-study heterogeneity. Begg’s test and Egger’ test were used to check for the publication bias. When the P-value was <0.05, publication bias was considered as significant. The aggregated sensitivity and specificity were observed by conducting the sensitivity analysis. All meta-analyses were performed using Stata software 12.0 (Stata Corporation, College Station, TX, USA).
Results
Characteristics of studies
Through searching PubMed, Embase, ISI Web of Knowledge, and Chinese National Knowledge Infrastructure databases, 25 publications were initially retrieved. Eight duplicate articles were removed. After reading titles and abstracts, four articles were excluded due to uncorrelated contents. Furthermore, full-text was read to eliminate irrelevant literatures, and two articles were eliminated because they had no sufficient data of RASSF1A promoter methylation. Finally, 10 publications with 422 patients and 219 controls were identified in this meta-analysis. Of these studies, seven studies were performed in Caucasians,30–36 while three studies were conducted in Asians.37–39 The detection of RASSF1A promoter methylation applied methylation-specific polymerase chain reaction and quantitative methylation-specific polymerase chain reaction. With regard to the type of control sample, normal tissue was used in five studies,31,33,35,36,39 four studies applied benign tissue,31,32,35,38 and adjacent tissue was applied in two studies34,37 (Figure 1; Tables 1 and S1–S6).
Figure 1.
Flow chart of study selection procedure.
Table 1.
Characteristics of eligible studies in the meta-analysis
| References (year) | Country | Ethnicity | Histology | Control
|
Tumor
|
Methods | Control | QA score | ||
|---|---|---|---|---|---|---|---|---|---|---|
| U (n) | M (n) | U (n) | M (n) | |||||||
| Santoro et al30 (2013) | Italy | Caucasian | TC | NR | NR | 10 | 9 | MSP | NR | NR |
| Brait et al31 (2012) | USA | Caucasian | TC | 1 | 13 | 5 | 38 | QMSP | NT | 6 |
| Brait et al31 (2012) | USA | Caucasian | TC | 1 | 43 | 5 | 38 | QMSP | BT | 6 |
| Qu and Xue37 (2012) | People’s Republic of China | Asian | TC | 21 | 7 | 13 | 15 | MSP | AT | 8 |
| Dai et al38 (2011) | People’s Republic of China | Asian | TC | 22 | 10 | 20 | 30 | MSP | BT | 8 |
| Mohammadi et al32 (2011) | Iran | Caucasian | TC | 2 | 23 | 6 | 19 | MSP | BT | 7 |
| Wang et al39 (2009) | People’s Republic of China | Asian | TC | 10 | 0 | 25 | 63 | MSP | NT | 6 |
| Lee et al33 (2008) | Sweden | Caucasian | FTC | 16 | 5 | 3 | 18 | MSP | NT | 7 |
| Nakamura et al34 (2005) | USA | Caucasian | TC | 27 | 0 | 51 | 27 | MSP | AT | 7 |
| Xing et al35 (2004) | USA | Caucasian | TC | 14 | 0 | 32 | 19 | QMSP | NT | 7 |
| Xing et al35 (2004) | USA | Caucasian | TC | 14 | 0 | 5 | 4 | QMSP | BT | 7 |
| Schagdarsurengin et al36 (2002) | Germany | Caucasian | TC | 3 | 1 | 11 | 27 | MSP | NT | 6 |
Abbreviations: AT, adjacent tissue; BT, benign tissue; FTC, follicular thyroid carcinoma; M, methylation sample; MSP, methylation-specific PCR; NR, not reported; NT, normal tissue; PCR, polymerase chain reaction; QA, quality assessment; QMSP, quantitative methylation-specific PCR; TC, thyroid carcinoma; U, unmethylation sample.
Quantitative data synthesis
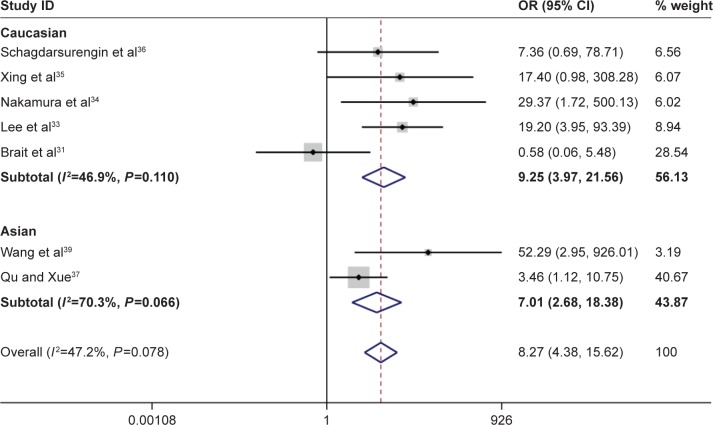
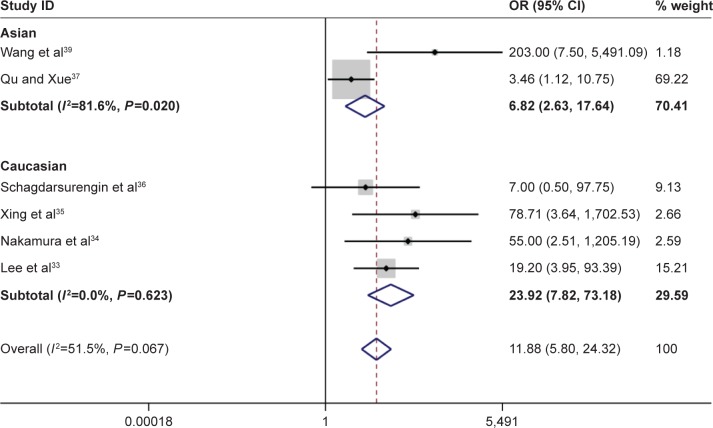
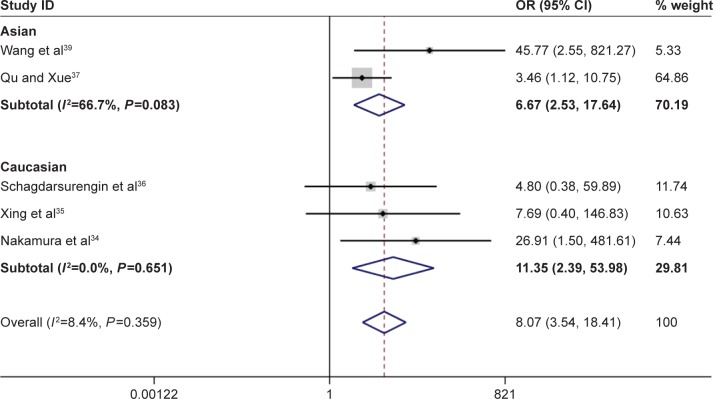
According to this meta-analysis, there was a significant association of RASSF1A promoter methylation with thyroid cancer risk (OR=5.94, 95% CI=2.09–16.89, P<0.05). When stratified by the ethnicity, a significant increased risk of thyroid cancer was detected in Asians, but not in Caucasians. In the stratified analysis by sample type, significantly increased risks were found in both normal tissues and adjacent tissues in detection of RASSF1A promoter methylation in thyroid cancer (normal tissue: OR=9.55, CI=4.21–21.67, P<0.05; adjacent tissue: OR=6.80, CI=2.49–18.56, P<0.05). Furthermore, significant association was also found in both FTC and PTC (for FTC: OR=11.88, CI=5.80–24.32, P<0.05; for PTC: OR=8.07, CI=3.54–18.41, P<0.05). In evaluating the association for pathological stage of thyroid cancer, no significant correlations of RASSF1A promoter methylation with distant metastasis and TNM-stage were observed (Figures 2–4; Table 2).
Figure 2.
Forest plot for the association between Ras-associated domain family 1A promoter methylation and risk of thyroid cancer.
Abbreviations: CI, confidence interval; OR, odds ratio.
Figure 3.
Forest plot for the association between Ras-associated domain family 1A promoter methylation and risk of follicular thyroid carcinoma.
Abbreviations: CI, confidence interval; OR, odds ratio.
Figure 4.
Forest plot for the association between Ras-associated domain family 1A promoter methylation and risk of papillary thyroid carcinoma.
Abbreviations: CI, confidence interval; OR, odds ratio.
Table 2.
Meta-analysis of association between Ras-associated domain family 1A promoter methylation and thyroid cancer risk
| Variables | Controls/cases (n) | OR | 95% CI | Heterogeneity
|
|
|---|---|---|---|---|---|
| I2 (%) | P-value | ||||
| Total | 118/347 | 8.27 | 4.38–15.62 | 47.20 | 0.078 |
| Ethnicity | |||||
| Caucasian | 80/231 | 9.25 | 3.97–21.56 | 46.90 | 0.110 |
| Asian | 38/116 | 7.01 | 2.68–18.38 | 70.30 | 0.066 |
| Disease type | |||||
| FTC | 104/94 | 11.88 | 5.80–24.32 | 51.50 | 0.067 |
| PTC | 83/166 | 8.07 | 3.54–18.41 | 8.4 | 0.359 |
| Sample type | |||||
| Normal tissue | 63/241 | 9.55 | 4.21–21.67 | 51.80 | 0.081 |
| Adjacent tissue | 55/106 | 6.80 | 2.49–18.56 | 58.10 | 0.122 |
| Pathogenesis | |||||
| Distant metastasis | 183/67 | 1.18 | 0.64–2.16 | 34.60 | 0.191 |
| TNM-stage | 64/32 | 1.79 | 0.72–4.44 | 52.60 | 0.121 |
Abbreviations: CI, confidence interval; FTC, follicular thyroid carcinoma; OR, odds ratio; PTC, papillary thyroid carcinoma.
Literature qualities
In this meta-analysis, 10 studies were scored by NOS table by two independent authors. The scores of eligible literatures ranged from 6 to 8, suggesting that the quality of included studies were of a high quality and the inclusion criteria were satisfied. The detailed information is shown in Table 1.
Publication bias and sensitivity analysis
The results of Begg’s test and Egger’s test indicated that no significant publication bias existed in the analysis of association between RASSF1A promoter methylation and thyroid cancer risk or pathological characteristics. (For risk of thyroid: Begg’s test P=0.453, Egger’s test P=0.377; for distant metastasis: Begg’s test P=0.624, Egger’s test P=0.669; for TNM-stage: Begg’s test P=0.117, Egger’s test P=0.441). Sensitivity analysis indicated that the results were stable in this meta-analysis, and the pooled ORs did not have a significant change by omitting each study (Figures 5–7).
Figure 5.
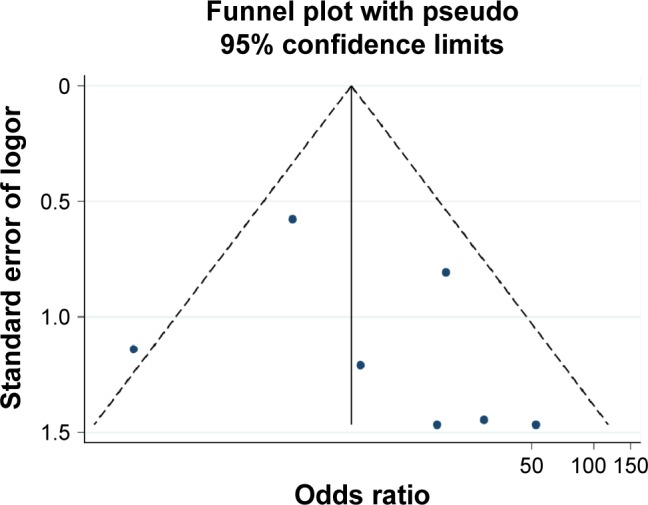
Funnel plot of publication biases on the association between Ras-associated domain family 1A promoter methylation and thyroid cancer risk.
Figure 6.
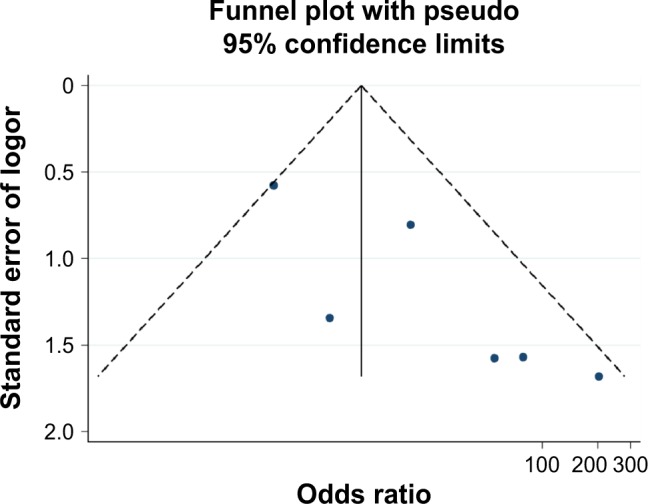
Funnel plot of publication biases on the association between Ras-associated domain family 1A promoter methylation and follicular thyroid carcinoma risk.
Figure 7.
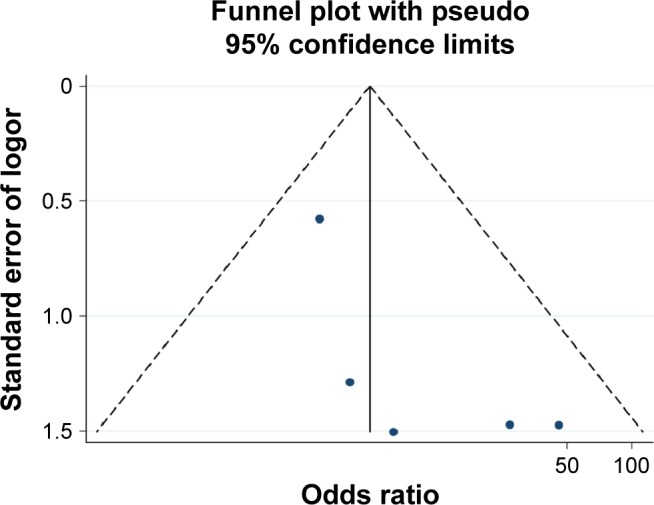
Funnel plot of publication biases on the association between Ras-associated domain family 1A promoter methylation and papillary thyroid carcinoma risk.
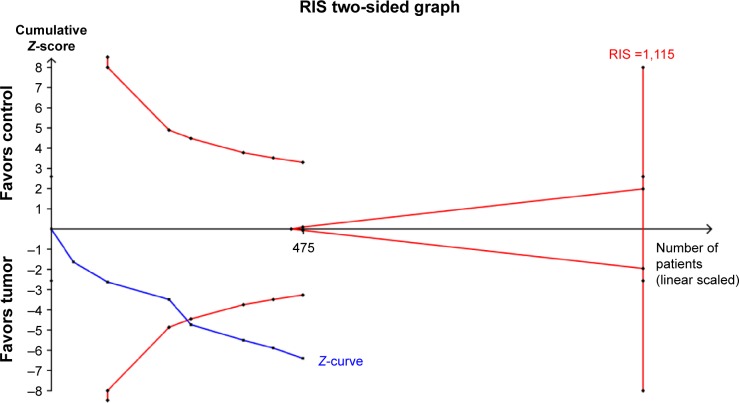
Trial sequential analysis
Trial sequential analysis indicated that Z-curve crossed the traditional boundary and the monitoring boundary for benefit, which indicated that no further studies were performed to study this association of RASSF1A promoter methylation with thyroid cancer risk. The Z-curve did not reach the boundary of RIS, and the RIS was 1,115. In other associations, no trial sequential analysis was conducted for the small number of samples (Figure 8).
Figure 8.
Trial sequential analysis comparing the Ras-associated domain family 1A promoter methylation frequency of control group and thyroid cancer group.
Abbreviation: RIS, required information size.
Discussion
Epigenetic alternations play an important role in the regulation of gene expression, especially in mammalian cells. DNA methylation usually occurs in CpG islands – clusters of CpGs of gene promoter region, which can lead to gene activation or inactivation. The hypermethylation of gene promoter represses gene expression by inhibiting gene transcription and further affects the total cell signal pathway. The inactivation of RASSF1A, one of tumor suppressor genes, is critical in the tumorigenesis of thyroid cancer. The changes in gene structure, deletion, or mutation can result in the irreversible loss of RASSF1A function. Alternatively, the cumulative RASSF1A methylation can also influence the expression and affect the progression of cancers, such as tumor differentiation and metastasis. In previous studies of thyroid cancers, many genes methylation have a significant association with the development of thyroid tumor. For example, p16INK4A/CDKN2A, p21CIP1/CDKN1B, and p27KIP1/CDKN1B, considering as tumor suppressors, can regulate the activity of cyclin–CDK complexes in mammalian cells. However, the expression of these genes in which promoter methylation is detected in 30% of thyroid tumor was commonly downregulated in thyroid cancer patients.40,41 In addition, the hypermethylation of thyrotropin receptor gene, receptor, fibroblast growth factor type 2, and receptor, fibroblast growth factor type 1 existed in thyroid cancers.42–44 RASSF1A protein, lacking enzymatic activity, contains a Ras-association domain and can regulate the cell cycle and apoptosis.13,14 Many studies have found that the RASSF1A promoter inactivation was frequent in thyroid cancers, >30% of thyroid tumors,33,34,37,39 but other reports also observed that the RASSF1A promoter methylation did not have significant correlations with thyroid cancer risk.31,35,36 Thus, it is necessary to conduct a meta-analysis to determine the strength of association between RASSF1A promoter methylation and thyroid cancer. In the literature retrieval, a previous meta-analysis was found.45 However, the meta-analysis only explored the association between RASSF1A promoter methylation and PTC risk, and no clinical information was included. The result of the previous study indicated a significant association of RASSF1A promoter methylation with PTC risk. This was the same result in this meta-analysis. To discuss the association of RASSF1A promoter methylation with FTC risk, distant metastasis, and TNM-stage, the meta-analysis was conducted.
In this study, the results indicated that aberrant methylation of RASSF1A promoter was more frequently detected in thyroid cancer than in noncancerous controls. In the included studies, although the results of Wang et al39 and Qu et al37 demonstrated that the frequency of RASSF1A promoter methylation increased the risk of thyroid cancer, the pooled ORs and 95% CI show that there was a significant association between RASSF1A promoter methylation and the risk of thyroid cancer in Asians. Furthermore, a small degree of heterogeneity among studies was noticed in the overall analysis. Therefore, the stratified analysis, on the basis of sample type and ethnicity, was carried out to reduce the heterogeneity. At the same time, the pooled ORs indicated that the significant association was found in the stratified analysis based on ethnicity and sample type. Furthermore, between-study heterogeneity, to a large extent, disappeared in the subgroup analysis. This phenomenon indicated that the sample type and ethnicity were the primary cause of heterogeneity. Interestingly, no significant association between the frequency of RASSF1A promoter methylation in thyroid malignant tumor and the frequency in thyroid benign tumor existed. In other subgroups, the higher frequency of FTC and PTC group was found, compared with the nontumorous group. From all eligible studies in this meta-analysis, no results indicated that the RASSF1A promoter methylation had a significant influence in the metastasis of thyroid tumor.30,36–39 In the meantime, the result of this statistical analysis was similar to the previous studies (for M0-stage and M1-stage: OR=1.18, 95% CI=0.64–2.16, P<0.05). Moreover, no significant association between the frequency of RASSF1A methylation and TNM-stage of thyroid cancer (for I-stage and II–IV-stage: OR=1.79, 95% CI=0.72–4.44, P<0.05) was found. In the included studies, one study found a significant association,38 but two studies obtained negative results.30,38 These results were inconsistent, but now, to a large extent, we could say that the frequency of RASSF1A promoter methylation might not be related with the TNM-stage of thyroid cancer. Certainly, the small sample size may limit the power of statistics. From the trial sequential analysis, although no studies were continuously performed to evaluate the association of RASSF1A promoter methylation with thyroid cancer risk, further studies, investigating the association between RASSF1A promoter methylation and thyroid cancer pathogenesis, were still needed to be carried out.
In this meta-analysis, the publication bias was not detected in the eligible studies. Begg’s test, Egger’s test, and funnel plot were performed, which indicated that the data did not have a considerable discrepancy among studies. Furthermore, consistent results were found in sensitivity analysis. However, several potential limitations must be emphasized in this study. First, with few included studies in this meta-analysis, the influence of small sample size of case and control cannot be ruled out. Second, the value of cut-off point of methylation cannot be achieved, thus the sensitivity and specificity of methylation about thyroid cancer risks cannot be determined. Third, the clinical information was so little that the analysis of association of RASSF1A promoter methylation with the pathogenesis of thyroid cancer patients cannot be performed.
Conclusion
This meta-analysis showed that RASSF1A promoter methylation might play an important role in thyroid cancer initiation and progression. The RASSF1A promoter methylation might be a promising biomarker for the early diagnosis of thyroid cancer. In addition, RASSF1A promoter methylation was associated with the increased risk of distant metastasis and late TNM-stage of thyroid cancer patients. However, in consideration of the limitations acknowledged above, more large-scale, multicenter, and well-designed case–control or cohort researches will provide more insights into the role of RASSF1A promoter methylation in the risk and pathogenesis of thyroid cancer.
Supplementary materials
Table S1.
Thyroid cancer risk
| References | Country | Ethnicity | Histology | U (n) | M (n) | U (n) | M (n) | Methods | Control type | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | NR | NR | 10 | 9 | MSP | NR | NR |
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 13 | 5 | 38 | QMSP | Normal tissue | 6 |
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 43 | 5 | 38 | QMSP | Benign tissue | 6 |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | Adjacent tissue | 8 |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 22 | 10 | 20 | 30 | MSP | Benign tissue | 8 |
| Mohammadi5 | Iran | Caucasian | Thyroid carcinoma | 2 | 23 | 6 | 19 | MSP | Benign tissue | 7 |
| Wang et al6 | China | Asian | Thyroid carcinoma | 10 | 0 | 25 | 63 | MSP | Normal tissue | 6 |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP | Normal tissue | 7 |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 51 | 27 | MSP | Adjacent tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 32 | 19 | QMSP | Normal tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 5 | 4 | QMSP | Benign tissue | 7 |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 11 | 27 | MSP | Normal tissue | 6 |
| Normal and adjacent tissue | ||||||||||
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 13 | 5 | 38 | QMSP | Normal tissue | 6 |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | Adjacent tissue | 8 |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 10 | 0 | 25 | 63 | MSP | Normal tissue | 6 |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP | Normal tissue | 7 |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 51 | 27 | MSP | Adjacent tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 32 | 19 | QMSP | Normal tissue | 7 |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 11 | 27 | MSP | Normal tissue | 6 |
| Benign tissue | ||||||||||
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 43 | 5 | 38 | QMSP | Benign tissue | 6 |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 22 | 10 | 20 | 30 | MSP | Benign tissue | 8 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 5 | 4 | QMSP | Benign tissue | 7 |
| Mohammadi et al5 | Iran | Caucasian | Thyroid carcinoma | 2 | 23 | 6 | 19 | MSP | Benign tissue | 7 |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; NOS, Newcastle–Ottawa scale; NR, not reported; PCR, polymerase chain reaction; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.
Table S2.
Meta-analysis table
| Variables | Controls/cases (n) | OR | 95% CI | I2 (%) | P-value |
|---|---|---|---|---|---|
| Heterogeneity | |||||
| Total | 136/365 | 5.94 | 2.09–16.89 | 55.40 | 0.028 |
| Ethnicity | |||||
| Caucasian | 98/249 | 5.4 | 1.40–20.86 | 59.60 | 0.03 |
| Asian | 38/116 | 10.02 | 0.63–159.62 | 70.30 | 0.066 |
| Disease type | |||||
| FTC | 104/94 | 11.88 | 5.80–24.32 | 51.50 | 0.067 |
| PTC | 152/133 | 2.18 | 0.48–9.83 | 68.40 | 0.007 |
| Sample type | |||||
| Normal tissue | 81/259 | 5.85 | 1.43–23.91 | 62.60 | 0.02 |
| Adjacent tissue | 55/106 | 7.29 | 0.77–69.05 | 58.10 | 0.122 |
| Pathogenesis | |||||
| Distant metastasis | 183/67 | 1.18 | 0.64–2.16 | 34.60 | 0.191 |
| TNM-stage | 64/32 | 1.79 | 0.72–4.44 | 52.60 | 0.121 |
Abbreviations: CI, confidence interval; FTC, follicular thyroid carcinoma; OR, odds ratio; PTC, papillary thyroid cancer.
Table S3.
TNM-stage
| References | Country | Ethnicity | Histology | Thyroid tumor
|
Methods | |||
|---|---|---|---|---|---|---|---|---|
| TNM-stage I
|
TNM-stage II–IV
|
|||||||
| U (n) | M (n) | U (n) | M (n) | |||||
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | 3 | 3 | 6 | 6 | MSP |
| Qu and Xue2 | People’s Republic of China | Asian | Thyroid carcinoma | 13 | 7 | 1 | 7 | MSP |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 15 | 23 | 5 | 7 | MSP |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; U, unmethylation sample.
Table S4.
Distant metastasis
| References | Country | Ethnicity | Histology | Thyroid tumor
|
Methods | |||
|---|---|---|---|---|---|---|---|---|
| M0 stage
|
M1 stage
|
|||||||
| U (n) | M (n) | U (n) | M (n) | |||||
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | 6 | 8 | 3 | 1 | MSP |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 11 | 8 | 3 | 6 | MSP |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 16 | 27 | 4 | 3 | MSP |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 30 | 43 | 3 | 13 | MSP |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 11 | 23 | 11 | 20 | MSP |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; U, unmethylation sample.
Table S5.
Papillary thyroid carcinoma
| References | Country | Ethnicity | Histology | Control
|
PTC
|
Methods | |||
|---|---|---|---|---|---|---|---|---|---|
| U (n) | M (n) | U (n) | M (n) | ||||||
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 10 | 0 | 19 | 42 | MSP | |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 23 | 11 | MSP | |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 24 | 6 | QMSP | |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 5 | 8 | MSP | |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; PTC, papillary thyroid cancer; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.
Table S6.
Follicular thyroid carcinoma
| References | Country | Ethnicity | Histology | Control
|
FTC
|
Methods | ||
|---|---|---|---|---|---|---|---|---|
| U (n) | M (n) | U (n) | M (n) | |||||
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 3 | 7 | MSP |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 3 | 9 | 14 | 0 | QMSP |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 1 | 14 | 10 | 0 | MSP |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 4 | 4 | 27 | 0 | MSP |
Abbreviations: FTC, follicular thyroid carcinoma; M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.
References
- 1.Santoro A, Pannone G, Carosi MA, et al. BRAF mutation and RASSF1A expression in thyroid carcinoma of southern Italy. J Cell Biochem. 2013;114(5):1174–1182. doi: 10.1002/jcb.24460. [DOI] [PubMed] [Google Scholar]
- 2.Brait M, Loyo M, Rosenbaum E, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics. 2012;7(7):710–719. doi: 10.4161/epi.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu F, Xue WJ. RASSF1A methylation and its clinical roles in papillary thyroid carcinoma. J Nantong University (Medical Sciences) 2012;32:490–491. [Google Scholar]
- 4.Dai YL, Cai DH, Chen H, Zhang Z, Zhang H, Li J. The association of the methylation of TSHR and RASSF1A with thyroid cancer. Shaanxi Med J. 2011;4011:1446–1449. [Google Scholar]
- 5.Mohammadi-asl J, Larijani B, Khorgami Z, et al. Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARbeta2 genes in papillary thyroid carcinoma. Med Oncol. 2011;28(4):1123–1128. doi: 10.1007/s12032-010-9587-z. [DOI] [PubMed] [Google Scholar]
- 6.Wang XH, Zhang GC, Liu YL, Sun GF, Nong WX, Yao ES. The detection of RASSF1A promoter methylation in thyroid tumor patients. Shaanxi Med J. 2009;38:790–792. [Google Scholar]
- 7.Lee JJ, Geli J, Larsson C, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol. 2008;33(4):861–869. [PubMed] [Google Scholar]
- 8.Nakamura N, Carney JA, Jin L, et al. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Lab Invest. 2005;85(9):1065–1075. doi: 10.1038/labinvest.3700306. [DOI] [PubMed] [Google Scholar]
- 9.Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64(5):1664–1668. doi: 10.1158/0008-5472.can-03-3242. [DOI] [PubMed] [Google Scholar]
- 10.Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002;62(13):3698–3701. [PubMed] [Google Scholar]
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212. doi: 10.1155/2013/965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito JP, Davies L. Is there really an increased incidence of thyroid cancer? Curr Opin Endocrinol Diabetes Obes. 2014;21(5):405–408. doi: 10.1097/MED.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 3.Enewold L, Zhu K, Ron E, et al. Rising thyroid cancer incidence in the United States by demographic and tumor characteristics, 1980–2005. Cancer Epidemiol Biomarkers Prev. 2009;18(3):784–791. doi: 10.1158/1055-9965.EPI-08-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris LG, Myssiorek D. Improved detection does not fully explain the rising incidence of well-differentiated thyroid cancer: a population-based analysis. Am J Surg. 2010;200(4):454–461. doi: 10.1016/j.amjsurg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elisei R, Pinchera A. Advances in the follow-up of differentiated or medullary thyroid cancer. Nat Rev Endocrinol. 2012;8(8):466–475. doi: 10.1038/nrendo.2012.38. [DOI] [PubMed] [Google Scholar]
- 6.Fagin JA, Wells SJ. Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375(11):1054–1067. doi: 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kebebew E, Ituarte PH, Siperstein AE, Duh QY, Clark OH. Medullary thyroid carcinoma clinical characteristics, treatment, prognostic factors, and a comparison of staging systems. Cancer. 2000;88(5):1139–1148. doi: 10.1002/(sici)1097-0142(20000301)88:5<1139::aid-cncr26>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Gilliland FD, Hunt WC, Morris DM, Key CR. Prognostic factors for thyroid carcinoma. A population-based study of 15,698 cases from the surveillance, epidemiology and end results (SEER) program 1973–1991. Cancer. 1997;79(3):564–573. doi: 10.1002/(sici)1097-0142(19970201)79:3<564::aid-cncr20>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Raue F, Kotzerke J, Reinwein D, et al. Prognostic factors in medullary thyroid carcinoma: evaluation of 741 patients from the German medullary thyroid carcinoma register. Clin Investig. 1993;71(1):7–12. doi: 10.1007/BF00210956. [DOI] [PubMed] [Google Scholar]
- 10.Roos L, van Dongen J, Bell CG, et al. Integrative DNA methylome analysis of pan-cancer biomarkers in cancer discordant monozygotic twin-pairs. Clin Epigenetics. 2016;8:7. doi: 10.1186/s13148-016-0172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kartal K, Onder S, Kosemehmetoglu K, Kilickap S, Tezel YG, Kaynaroglu V. Methylation status of TSHr in well-differentiated thyroid cancer by using cytologic material. BMC Cancer. 2015;15:824. doi: 10.1186/s12885-015-1861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis RJ, Wang Y, Stevenson HS, et al. Genome-wide methylation patterns in papillary thyroid cancer are distinct based on histological subtype and tumor genotype. J Clin Endocrinol Metab. 2014;99(2):E329–E337. doi: 10.1210/jc.2013-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen NP, Donninger H, Vos MD, et al. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007;26(42):6203–6211. doi: 10.1038/sj.onc.1210440. [DOI] [PubMed] [Google Scholar]
- 14.Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23(1–2):73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17(8):700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 16.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776(1):58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu GS, Lai CY, Xu Y, Bu CF, Su ZX. Aberrant methylation of RASSF1A gene contribute to the risk of renal cell carcinoma: a meta-analysis. Asian Pac J Cancer Prev. 2015;16(11):4665–4669. doi: 10.7314/apjcp.2015.16.11.4665. [DOI] [PubMed] [Google Scholar]
- 18.Huang YZ, Wu W, Wu K, Xu XN, Tang WR. Association of RASSF1A promoter methylation with lung cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2014;15(23):10325–10328. doi: 10.7314/apjcp.2014.15.23.10325. [DOI] [PubMed] [Google Scholar]
- 19.Jiang D, Shen Y, Dai D, et al. Meta-analyses of methylation markers for prostate cancer. Tumour Biol. 2014;35(10):10449–10455. doi: 10.1007/s13277-014-2300-7. [DOI] [PubMed] [Google Scholar]
- 20.Si JG, Su YY, Han YH, Chen RH. Role of RASSF1A promoter methylation in the pathogenesis of ovarian cancer: a meta-analysis. Genet Test Mol Biomarkers. 2014;18(6):394–402. doi: 10.1089/gtmb.2014.0022. [DOI] [PubMed] [Google Scholar]
- 21.Ashktorab H, Rahi H, Wansley D, et al. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8(8):807–815. doi: 10.4161/epi.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YS, Xie Q, Yang DY, Zheng Y. Role of RASSF1A promoter methylation in the pathogenesis of hepatocellular carcinoma: a meta-analysis of 21 cohort studies. Mol Biol Rep. 2014;41(6):3925–3933. doi: 10.1007/s11033-014-3260-0. [DOI] [PubMed] [Google Scholar]
- 23.Shi DT, Han M, Gao N, Tian W, Chen W. Association of RASSF1A promoter methylation with gastric cancer risk: a meta-analysis. Tumour Biol. 2014;35(2):943–948. doi: 10.1007/s13277-013-1123-2. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, Cui L, Chen WD, Shen SH, Ding LD. The prognostic role of RASSF1A promoter methylation in breast cancer: a meta-analysis of published data. PLoS One. 2012;7(5):e36780. doi: 10.1371/journal.pone.0036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19(4):251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 29.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 30.Santoro A, Pannone G, Carosi MA, et al. BRAF mutation and RASSF1A expression in thyroid carcinoma of southern Italy. J Cell Biochem. 2013;114(5):1174–1182. doi: 10.1002/jcb.24460. [DOI] [PubMed] [Google Scholar]
- 31.Brait M, Loyo M, Rosenbaum E, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics. 2012;7(7):710–719. doi: 10.4161/epi.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi-asl J, Larijani B, Khorgami Z, et al. Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARbeta2 genes in papillary thyroid carcinoma. Med Oncol. 2011;28(4):1123–1128. doi: 10.1007/s12032-010-9587-z. [DOI] [PubMed] [Google Scholar]
- 33.Lee JJ, Geli J, Larsson C, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol. 2008;33(4):861–869. [PubMed] [Google Scholar]
- 34.Nakamura N, Carney JA, Jin L, et al. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Lab Invest. 2005;85(9):1065–1075. doi: 10.1038/labinvest.3700306. [DOI] [PubMed] [Google Scholar]
- 35.Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64(5):1664–1668. doi: 10.1158/0008-5472.can-03-3242. [DOI] [PubMed] [Google Scholar]
- 36.Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002;62(13):3698–3701. [PubMed] [Google Scholar]
- 37.Qu F, Xue WJ. RASSF1A methylation and its clinical roles in papillary thyroid carcinoma. J Nantong University (Medical Sciences) 2012;32:490–491. [Google Scholar]
- 38.Dai YL, Cai DH, Chen H, Zhang Z, Zhang H, Li J. The association of the methylation of TSHR and RASSF1A with thyroid cancer. Shaanxi Med J. 2011;4011:1446–1449. [Google Scholar]
- 39.Wang XH, Zhang GC, Liu YL, Sun GF, Nong WX, Yao ES. The detection of RASSF1A promoter methylation in thyroid tumor patients. Shaanxi Med J. 2009;38:790–792. [Google Scholar]
- 40.Khoo ML, Beasley NJ, Ezzat S, Freeman JL, Asa SL. Overexpression of cyclin D1 and underexpression of p27 predict lymph node metastases in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2002;87(4):1814–1818. doi: 10.1210/jcem.87.4.8353. [DOI] [PubMed] [Google Scholar]
- 41.Elisei R, Shiohara M, Koeffler HP, Fagin JA. Genetic and epigenetic alterations of the cyclin-dependent kinase inhibitors p15INK4b and p16INK4a in human thyroid carcinoma cell lines and primary thyroid carcinomas. Cancer. 1998;83(10):2185–2193. [PubMed] [Google Scholar]
- 42.Khaled H, Al Lahloubi N, Rashad N. A review on thyroid cancer during pregnancy: multitasking is required. J Adv Res. 2016;7(4):565–570. doi: 10.1016/j.jare.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St BR, Zheng L, Liu W, Winer D, Asa SL, Ezzat S. Fibroblast growth factor receptors as molecular targets in thyroid carcinoma. Endocrinology. 2005;146(3):1145–1153. doi: 10.1210/en.2004-1134. [DOI] [PubMed] [Google Scholar]
- 44.Kondo T, Zheng L, Liu W, Kurebayashi J, Asa SL, Ezzat S. Epigenetically controlled fibroblast growth factor receptor 2 signaling imposes on the RAS/BRAF/mitogen-activated protein kinase pathway to modulate thyroid cancer progression. Cancer Res. 2007;67(11):5461–5470. doi: 10.1158/0008-5472.CAN-06-4477. [DOI] [PubMed] [Google Scholar]
- 45.Jiang JL, Tian GL, Chen SJ, Xu LI, Wang HQ. Promoter methylation of p16 and RASSF1A genes may contribute to the risk of papillary thyroid cancer: a meta-analysis. Exp Ther Med. 2015;10(4):1549–1555. doi: 10.3892/etm.2015.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Thyroid cancer risk
| References | Country | Ethnicity | Histology | U (n) | M (n) | U (n) | M (n) | Methods | Control type | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | NR | NR | 10 | 9 | MSP | NR | NR |
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 13 | 5 | 38 | QMSP | Normal tissue | 6 |
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 43 | 5 | 38 | QMSP | Benign tissue | 6 |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | Adjacent tissue | 8 |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 22 | 10 | 20 | 30 | MSP | Benign tissue | 8 |
| Mohammadi5 | Iran | Caucasian | Thyroid carcinoma | 2 | 23 | 6 | 19 | MSP | Benign tissue | 7 |
| Wang et al6 | China | Asian | Thyroid carcinoma | 10 | 0 | 25 | 63 | MSP | Normal tissue | 6 |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP | Normal tissue | 7 |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 51 | 27 | MSP | Adjacent tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 32 | 19 | QMSP | Normal tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 5 | 4 | QMSP | Benign tissue | 7 |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 11 | 27 | MSP | Normal tissue | 6 |
| Normal and adjacent tissue | ||||||||||
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 13 | 5 | 38 | QMSP | Normal tissue | 6 |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | Adjacent tissue | 8 |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 10 | 0 | 25 | 63 | MSP | Normal tissue | 6 |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP | Normal tissue | 7 |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 51 | 27 | MSP | Adjacent tissue | 7 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 32 | 19 | QMSP | Normal tissue | 7 |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 11 | 27 | MSP | Normal tissue | 6 |
| Benign tissue | ||||||||||
| Brait et al2 | USA | Caucasian | Thyroid carcinoma | 1 | 43 | 5 | 38 | QMSP | Benign tissue | 6 |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 22 | 10 | 20 | 30 | MSP | Benign tissue | 8 |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 5 | 4 | QMSP | Benign tissue | 7 |
| Mohammadi et al5 | Iran | Caucasian | Thyroid carcinoma | 2 | 23 | 6 | 19 | MSP | Benign tissue | 7 |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; NOS, Newcastle–Ottawa scale; NR, not reported; PCR, polymerase chain reaction; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.
Table S2.
Meta-analysis table
| Variables | Controls/cases (n) | OR | 95% CI | I2 (%) | P-value |
|---|---|---|---|---|---|
| Heterogeneity | |||||
| Total | 136/365 | 5.94 | 2.09–16.89 | 55.40 | 0.028 |
| Ethnicity | |||||
| Caucasian | 98/249 | 5.4 | 1.40–20.86 | 59.60 | 0.03 |
| Asian | 38/116 | 10.02 | 0.63–159.62 | 70.30 | 0.066 |
| Disease type | |||||
| FTC | 104/94 | 11.88 | 5.80–24.32 | 51.50 | 0.067 |
| PTC | 152/133 | 2.18 | 0.48–9.83 | 68.40 | 0.007 |
| Sample type | |||||
| Normal tissue | 81/259 | 5.85 | 1.43–23.91 | 62.60 | 0.02 |
| Adjacent tissue | 55/106 | 7.29 | 0.77–69.05 | 58.10 | 0.122 |
| Pathogenesis | |||||
| Distant metastasis | 183/67 | 1.18 | 0.64–2.16 | 34.60 | 0.191 |
| TNM-stage | 64/32 | 1.79 | 0.72–4.44 | 52.60 | 0.121 |
Abbreviations: CI, confidence interval; FTC, follicular thyroid carcinoma; OR, odds ratio; PTC, papillary thyroid cancer.
Table S3.
TNM-stage
| References | Country | Ethnicity | Histology | Thyroid tumor
|
Methods | |||
|---|---|---|---|---|---|---|---|---|
| TNM-stage I
|
TNM-stage II–IV
|
|||||||
| U (n) | M (n) | U (n) | M (n) | |||||
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | 3 | 3 | 6 | 6 | MSP |
| Qu and Xue2 | People’s Republic of China | Asian | Thyroid carcinoma | 13 | 7 | 1 | 7 | MSP |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 15 | 23 | 5 | 7 | MSP |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; U, unmethylation sample.
Table S4.
Distant metastasis
| References | Country | Ethnicity | Histology | Thyroid tumor
|
Methods | |||
|---|---|---|---|---|---|---|---|---|
| M0 stage
|
M1 stage
|
|||||||
| U (n) | M (n) | U (n) | M (n) | |||||
| Santoro et al1 | Italy | Caucasian | Thyroid carcinoma | 6 | 8 | 3 | 1 | MSP |
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 11 | 8 | 3 | 6 | MSP |
| Dai et al4 | People’s Republic of China | Asian | Thyroid carcinoma | 16 | 27 | 4 | 3 | MSP |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 30 | 43 | 3 | 13 | MSP |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 11 | 23 | 11 | 20 | MSP |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; U, unmethylation sample.
Table S5.
Papillary thyroid carcinoma
| References | Country | Ethnicity | Histology | Control
|
PTC
|
Methods | |||
|---|---|---|---|---|---|---|---|---|---|
| U (n) | M (n) | U (n) | M (n) | ||||||
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP | |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 10 | 0 | 19 | 42 | MSP | |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 27 | 0 | 23 | 11 | MSP | |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 14 | 0 | 24 | 6 | QMSP | |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 5 | 8 | MSP | |
Abbreviations: M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; PTC, papillary thyroid cancer; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.
Table S6.
Follicular thyroid carcinoma
| References | Country | Ethnicity | Histology | Control
|
FTC
|
Methods | ||
|---|---|---|---|---|---|---|---|---|
| U (n) | M (n) | U (n) | M (n) | |||||
| Qu and Xue3 | People’s Republic of China | Asian | Thyroid carcinoma | 21 | 7 | 13 | 15 | MSP |
| Lee et al7 | Sweden | Caucasian | Follicular thyroid carcinoma | 16 | 5 | 3 | 18 | MSP |
| Schagdarsurengin et al10 | Germany | Caucasian | Thyroid carcinoma | 3 | 1 | 3 | 7 | MSP |
| Xing et al9 | USA | Caucasian | Thyroid carcinoma | 3 | 9 | 14 | 0 | QMSP |
| Wang et al6 | People’s Republic of China | Asian | Thyroid carcinoma | 1 | 14 | 10 | 0 | MSP |
| Nakamura et al8 | USA | Caucasian | Thyroid carcinoma | 4 | 4 | 27 | 0 | MSP |
Abbreviations: FTC, follicular thyroid carcinoma; M, methylation sample; MSP, methylation-specific PCR; PCR, polymerase chain reaction; QMSP, quantitative methylation-specific PCR; U, unmethylation sample.