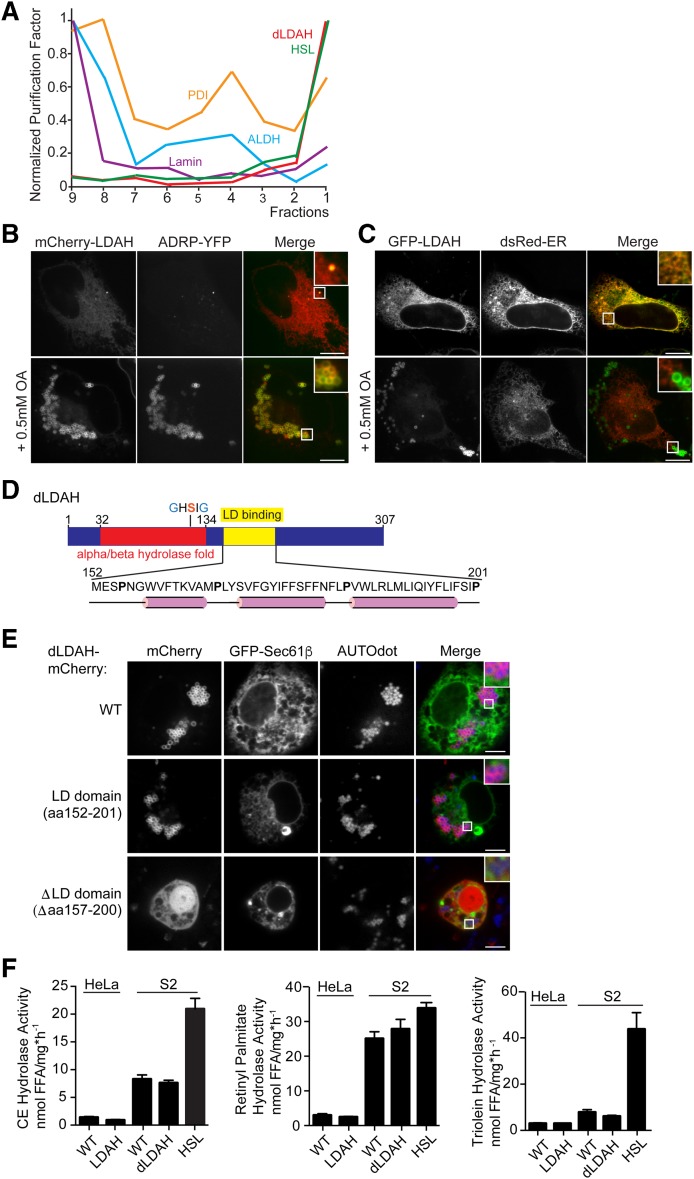
Fig. 1.
LDAH homologs localize to LDs with a hydrophobic hairpin. A: The Drosophila homolog of LDAH has the purification profile of a LD protein. Normalized purification factors of different organelle markers across a cellular fractionation are plotted. HSL is the LD marker; protein disulfide isomerase is the ER marker; alcohol dehydrogenase is the cytosolic marker; lamin is the nuclear marker. B, C: LDAH localizes to LDs or to the ER in the absence of LDs. B: mCherry-tagged LDAH colocalizes with ADRP in HeLa cells in the presence of LDs. Cells were transfected with constructs and treated with 0.5 mM oleic acid overnight. Representative images are shown. Scale bar, 10 μm. C: GFP-tagged LDAH localized to the ER in the absence of LDs. Cells were transfected with constructs and imaged the next day. Representative images are shown. Scale bar, 10 μm. D, E: A hydrophobic hairpin motif localizes LDAH to LDs. D: A hydrophobic segment comprising amino acids 160–195 of Drosophila CG9186/LDAH is predicted to have a hairpin structure and is responsible for LD binding. The α/β-hydrolase fold and the catalytic GxSxG-motif are indicated. α-Helices predicted by PSIPRED and JPred 4 are shown in pink. E: Full-length mCherry-tagged dLDAH and amino acids 152–201 of dLDAH localize to LDs after oleic acid treatment, while deletion of amino acids 157–200 results in ablation of LD binding. GFP-Sec61β was used to visualize the ER. Cells were transfected with constructs and treated with 1 mM oleic acid overnight. LDs were stained with AUTOdot (blue). Representative images are shown. Scale bar, 5 μm. F: LDAH overexpression does not affect cholesterol esterase, retinol esterase, or triacylglycerol hydrolysis activity. Nanomoles of free fatty acids (FFA) per (hour per milligram protein) ± SD. Values are means (n = 4). Activities were determined in lysates of WT HeLa or S2 cells and cells overexpressing the LDAH homologs using phospholipid-emulsified 3H-labeled lipids at neutral pH. S2 cells overexpressing HSL were used as a positive control. ALDH, alcohol dehydrogenase; PDI, protein disulfide isomerase