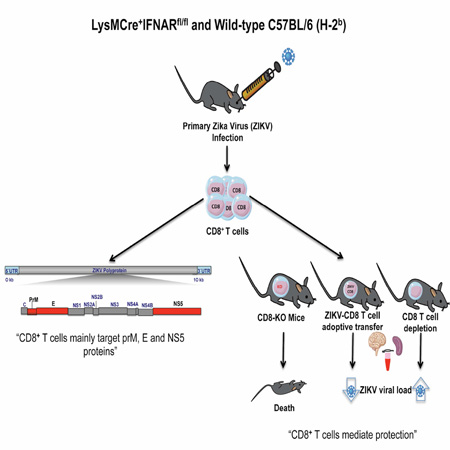
SUMMARY
CD8+ T cells protect against and promote the pathogenesis of some flaviviruses, but their role in Zika virus (ZIKV) infection is unknown. We developed mice lacking the type I interferon receptor specifically in a subset of myeloid cells (LysMCre+IFNARfl/fl C57BL/6 (H-2b) mice) as a model for ZIKV infection and used it to evaluate CD8+ T cell responses to ZIKV. 26 and 15 CD8+ T cell-reactive peptides for ZIKV African (MR766) and Asian (FSS13025) lineage strains respectively were identified and validated. CD8+ T cells from infected mice were polyfunctional and mediated cytotoxicity. Adoptive transfer of ZIKV-immune CD8+ T cells reduced viral burdens, whereas their depletion led to higher tissue burdens and CD8−/− mice displayed higher mortality to ZIKV-infection. Collectively, these results demonstrate that CD8+ T cells protect against ZIKV infection. Further, this study provides an immunocompetent mouse model for investigating ZIKV-specific T cell responses.
Keywords: Zika virus, CD8+ T cell, epitope, peptide, mouse model, LysMCre+IFNARfl/fl
Graphical abstract
eTOC Blurb
The nature and role of CD8+ T cell responses during Zika virus (ZIKV) infection is unknown Elong Ngono et al. develop a mouse model to identify, characterize and validate H-2b-restricted CD8+ T cell response to ZIKV and demonstrate a protective role for CD8+ T cells during primary ZIKV infection.
INTRODUCTION
Zika virus (ZIKV) is a flavivirus transmitted by Aedes species mosquitoes. Three genetic lineages have been identified: East African, West African, and Asian (Lanciotti et al., 2016). ZIKV strain MR766 of the East African lineage was isolated in the 1940s, whereas both West African and Asian strains were discovered in the 1960s. Identification and diagnosis of ZIKV has been and continues to be confounded by its overlap in geographic range, vector space, symptomology and serological cross-reactivity with other flaviviruses such as dengue virus (DENV) (Ioos et al., 2014; Zammarchi et al., 2015).
A large body of literature has provided evidence for a potential dual role for CD8+ T cells in protection and pathogenesis during DENV infection (Screaton et al., 2015; Tang et al., 2015; Weiskopf and Sette, 2014; Zellweger and Shresta, 2014). Epidemiologic studies indicate that Severe Dengue is most often seen in individuals experiencing a heterotypic DENV infection after prior seroconversion to at least one of the other three serotypes (Guzman et al., 2000; Sangkawibha et al., 1984). Some studies showed cross-reactive CD8 T cells are more activated during secondary infection (Mongkolsapaya et al., 2003) with a suboptimal T cell phenotype (Mongkolsapaya et al., 2006) (Imrie et al., 2007; Mangada and Rothman, 2005) suggesting a possible pathogenic role for cross-reactive T cells. However, recently emerging literature points to a protective role for T cells in DENV infection (Weiskopf et al., 2013; Weiskopf et al., 2015), and our previous work on DENV using mouse models (Prestwood et al., 2012b; Yauch et al., 2010; Yauch et al., 2009; Zellweger et al., 2014; Zellweger et al., 2013; Zellweger et al., 2015) in C57BL/6 and 129/Sv mice lacking type I IFN receptor (IFNAR) alone or both type I and II IFN receptors (AB6, A129, and AG129) has provided multiple lines of evidence indicating a protective role for CD8+ T cells.
H-2b mouse models of ZIKV infection recently have been established in WT C57BL/6 mice treated with blocking anti-IFNAR monoclonal antibody and in gene-deficient mice that globally lack IFNAR or both IFNAR and type II IFN receptors (Dowall et al., 2016; Govero et al., 2016; Lazear et al., 2016; Rossi et al., 2016). To investigate IFN receptor-competent CD8+ T cell responses in H-2b mice, in the present study we established a model of ZIKV infection in LysMCre+IFNARfl/fl C57BL/6 mice, which lack IFNAR in a subset of myeloid cells but express normal IFNAR levels on T cells, B cells, and most dendritic cells (Clausen et al., 1999; Diamond et al., 2011). We infected both LysMCre+IFNARfl/fl C7BL/6 mice and anti-IFNAR antibody-treated wild-type (WT) C57BL/6 mice with ZIKV MR766 and FSS13025 strains and mapped the H-2b-restricted CD8+ T cell responses. Additionally, we demonstrated a protective role for CD8+ T cells in controlling ZIKV infection in LysMCre+IFNARfl/fl mice. Our work provides an immunocompetent and well-characterized H-2b mouse model for investigating protective vs. pathogenic roles of ZIKV-specific CD8+ T cell responses.
RESULTS
Characterization of CD8+ T cell response in WT C57BL/6 mice treated with IFNAR-blocking antibody
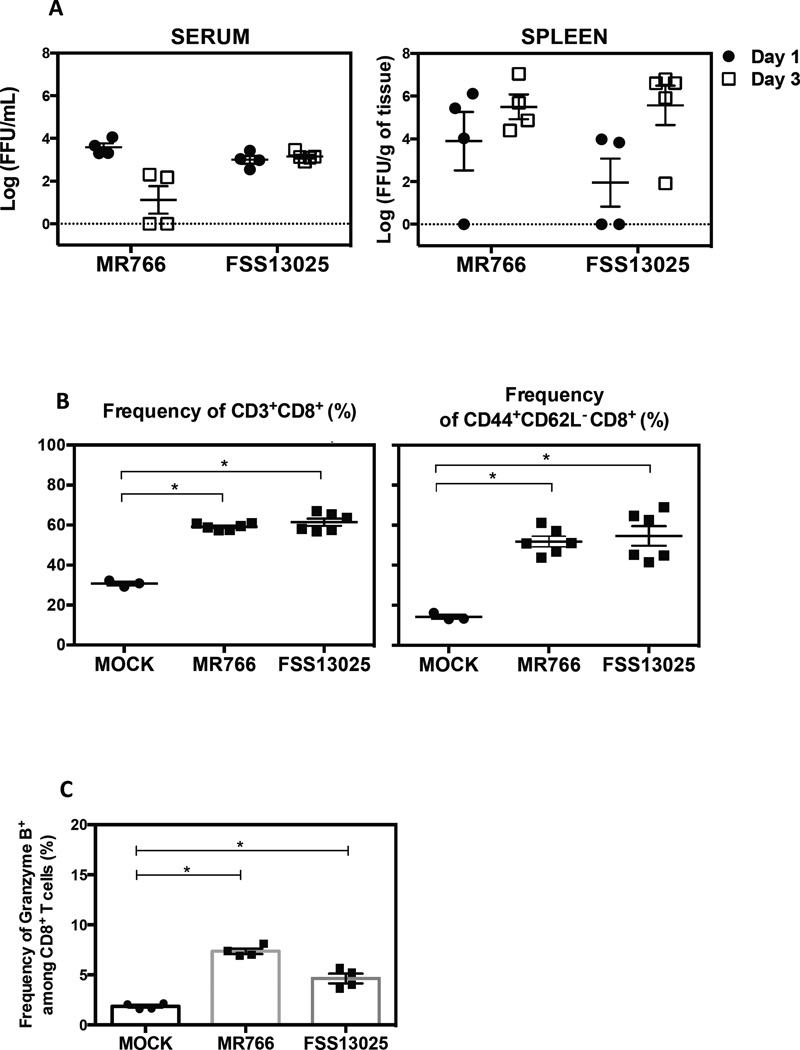
H-2b mice that are genetically deficient in IFNAR or treated with IFNAR-blocking antibody are susceptible to ZIKV infection (Dowall et al., 2016; Lazear et al., 2016; Miner et al., 2016; Rossi et al., 2016). To characterize the CD8+ T cell response in H-2b mice, WT C57BL/6 mice were treated with an IFNAR-blocking antibody MAR1-5A3 (Sheehan et al., 2006) prior to inoculation with ZIKV strains MR766 or FSS13025 and infectious virus particles in serum and spleen were measured at days 1 and 3 post-infection. At day 1, infectious ZIKV was detectable in all of the sera and some spleens from mice treated with anti-IFNAR antibody (Figure 1A). Three days after infection, the viral load decreased in serum of mice infected with MR766 but not those infected with FSS13025. In the spleen, the level of infectious virus in both groups increased at day 3 compared to day 1.
Figure 1. Characterization of ZIKV infection and identification of epitopes recognized by CD8+ T cells in WT C57BL/6 mice treated with IFNAR-blocking antibody.
WT C57BL/6 mice were administered IFNAR-blocking antibody (MAR1-5A3) one day prior to infection with 104 Focus Forming Units (FFU) of ZIKV strains MR766 (n=4) or FSS13025 (n=4). MOCK represents control mice injected with 10% FBS/PBS. (A) Levels of infectious ZIKV in serum and spleen on day 1 or 3 post-infection were measured by BHK-21 cell-based FFA. (B) The expansion of total CD8+ T cells and CD44+CD62L− CD8+ T cells in MOCK group (n=3) or ZIKV-infected mice (MR766 and FSS13025, n=4) was determined on day 7 post-infection. (C) The percentage of granzyme B produced by infected (n=4) and MOCK mice (n=4) is represented. Kruskall Wallis test was used first to compare all groups following by Mann-Whitney test to compare MOCK vs. each ZIKV-infected mouse group. See also Figure S1.
Having confirmed replication of both ZIKV strains in this mouse model, we next assessed the CD8+ T cell response in the spleen 7 days after infection. The frequencies of total CD8+ T cells and antigen-experienced (CD44+CD62L−) CD8+ T cells were increased in infected mice with IFNAR blockade relative to MOCK mice (Figure 1B). Infected mice contained 2-fold more total or antigen-experienced CD8+ T cells than MOCK mice. In addition, ZIKV MR766- and FSS13025-infected mice, respectively, contained 5- and 3-fold more CD8+ T cells expressing granzyme B (a marker of cytotoxicity) compared to controls (Figure 1C). No difference in CD8+ T cell response was observed between MR766 and FSS13025 groups. These results indicate that ZIKV induces CD8+ T cell expansion and activation in WT mice treated with IFNAR-blocking antibody, and that this model is suitable for identifying ZIKV-derived epitopes recognized by CD8+ T cells.
Identification of ZIKV-derived epitopes recognized by CD8+ T cells from WT C57BL/6 mice treated with IFNAR-blocking antibody
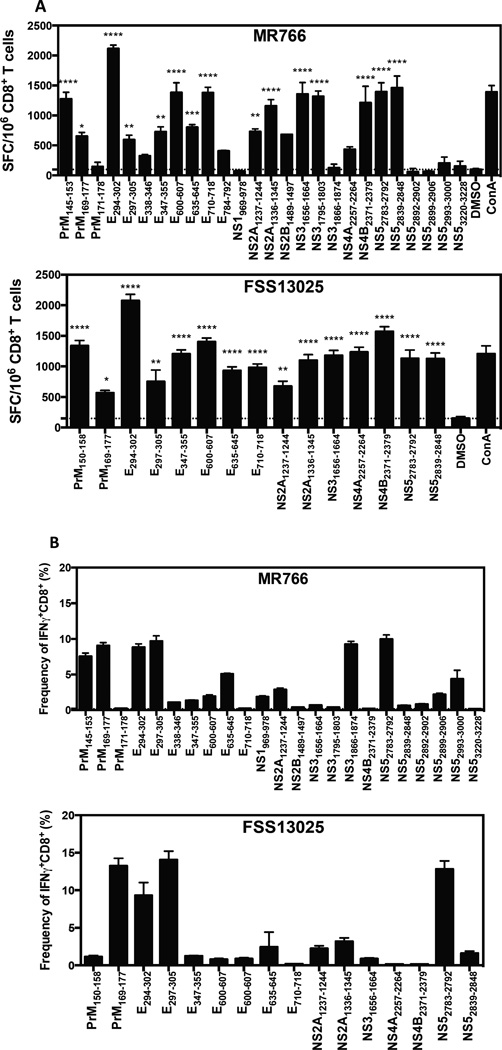
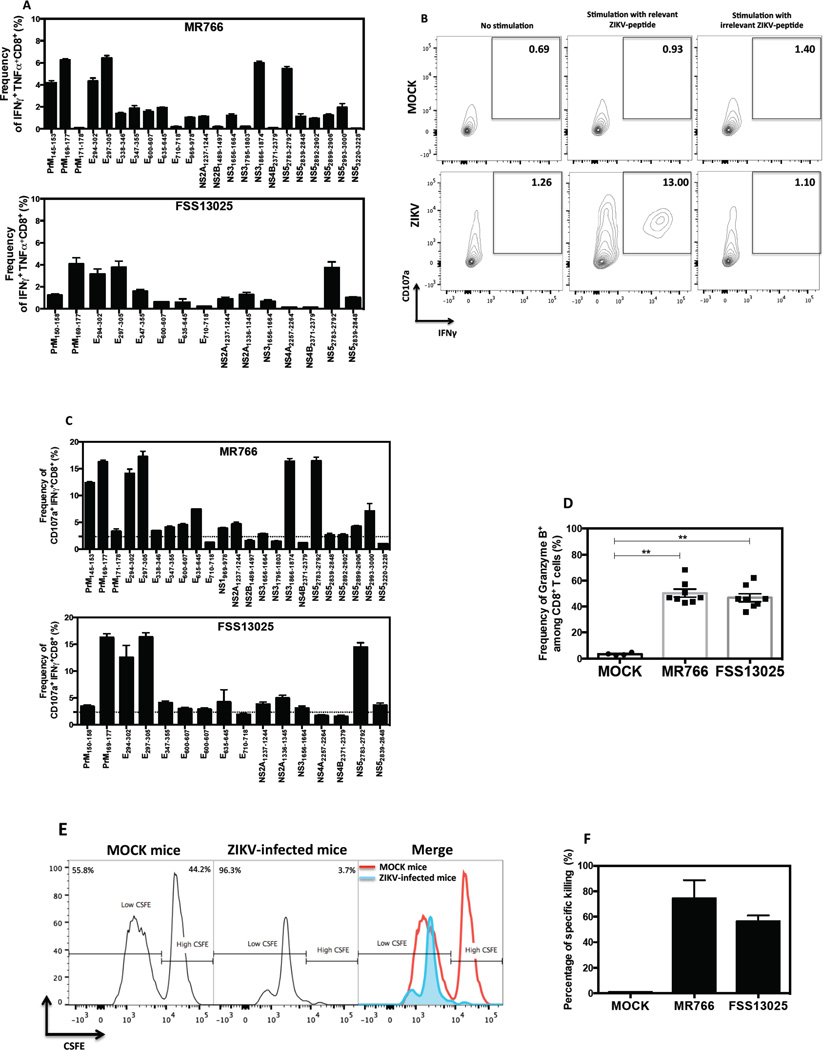
To map the specificity of the MHC class I–restricted CD8+ T cell response, we first inspected the proteome of ZIKV for the presence of peptides predicted to bind H-2b class I molecules (Kb and Db) with high affinity using a bioinformatic prediction program (Kim et al., 2012). A total of 244 predicted H-2b-binding peptides were identified with 202 shared between both ZIKV strains, and 42 specific for FSS13025. Among these peptides, 96 were specific for H-2Kb, 148 for H-2Db, and 22 were predicted to bind both MHC class I alleles. We next tested all peptides individually in an IFNγ-ELISPOT assay using CD8+ T cells from mice infected with either MR766 or FSS13025. Twenty-six peptides were positive for ZIKV MR766 (Supplementary Figure 1A, top panel) and 15 peptides were positive for ZIKV FSS13025 (Supplementary Figure 1A, bottom panel).
The identified ZIKV epitopes are derived from 9 (for MR766) and from 7 (for FSS13025) of the 10 ZIKV proteins including structural proteins prM and E, and non-structural proteins NS1, NS2B (only MR766), NS2A, NS3, NS4A, NS4B and NS5 (Supplementary Figure 1A). Although the CD8+ T cell responses to ZIKV MR766 and ZIKV FSS13025 were not identical, the E protein-derived epitopes predominated in both ZIKV strains.
To validate the identification of these ZIKV-derived peptides, intracellular cytokine staining (ICS) was performed. Splenocytes were stimulated with all positive peptides and the frequency of IFNγ-producing CD8+ T cells was reported for each peptide for both ZIKV strains (Supplementary Figure 1B). Five of 26 MR766-derived peptides (top panel) and four of 15 FSS13025-derived peptides (bottom panel) induced a high frequency of IFNγ-expressing cells. The following 4 peptides were shared between both ZIKV strains: prM169–177, E294–302, E297–305 and NS52783–2792. Only NS31886–1874 was specific for ZIKV MR766. For both ZIKV strains, all positive peptides confirmed by IFNγ-ICS induced a high frequency of IFNγ+TNFα+ and CD107a+IFNγ+ double-positive cells (Supplementary Figure 1 C–D). CD8+ T cells from mice infected with both ZIKV strains exhibited cytolytic activity by killing approximately 70% of splenocytes loaded with prM169–177, E297–305 and NS52783–2792 peptides (Supplementary Figure 1E). These results demonstrate a polyfunctional phenotype of ZIKV antigen-specific CD8+ T cells upon infection with either ZIKV strain in C57BL/6 mice after IFNAR blockade.
LysMCre+IFNARfl/fl mice, a H-2b model susceptible to ZIKV infection
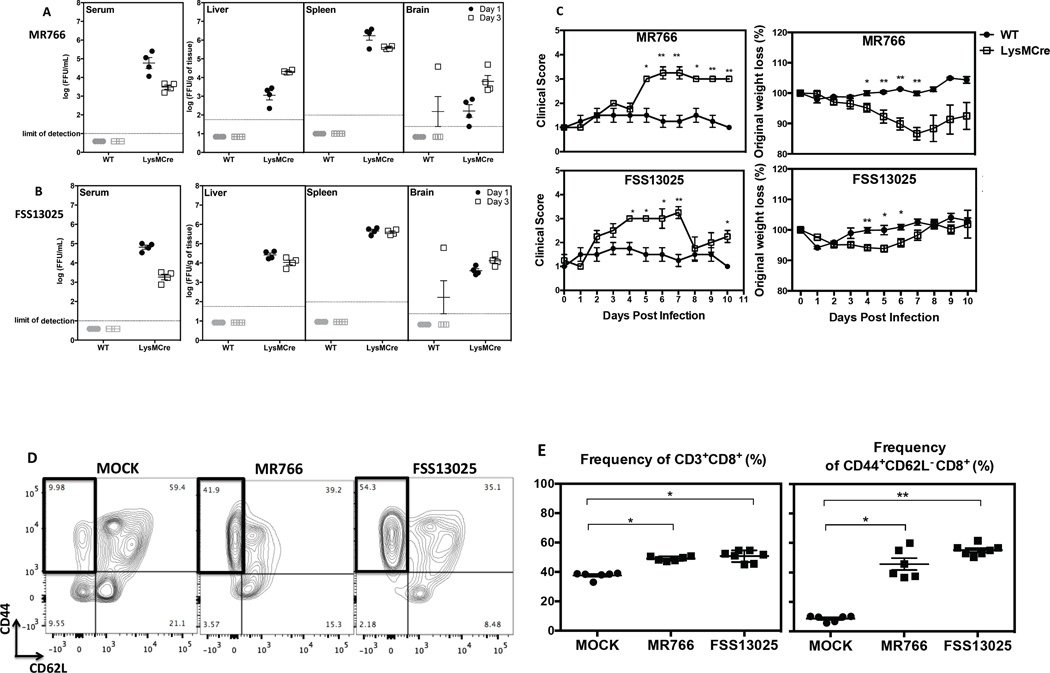
To investigate the role of CD8+ T cells during ZIKV infection in a more immunocompetent model than mice with global IFNAR blockade, LysMCre+IFNARfl/fl C57BL/6 mice, recently published for utility in studying DENV infection (Pinto et al., 2015), were evaluated; these mice display normal T and B cell immune responses and lack IFNAR expression only in a subset of myeloid cells. The Ifnar gene deletion is efficient in mature macrophages (83–98%) and granulocytes (100%) but partial for CD11C+ splenic dendritic cells (16%) (Clausen et al., 1999; Diamond et al., 2011). LysMCre+IFNARfl/fl and WT C57BL/6 mice were infected intravenously with MR766 or FSS13025, and levels of infectious virus in serum, liver, spleen, and brain at 1 and 3 days after infection were determined. At day 1 post-infection, the infectious virus was detectable in all of the tissues tested in LysMCre+IFNARfl/fl mice infected with MR766 (Figure 2A) and FSS13025 (Figure 2B), whereas virus was undetectable in WT mice. At day 3 post-infection, infectious ZIKV were still detectable in tissues of LysMCre+IFNARfl/fl mice. Based on these results, LysMCre+IFNAR1fl/fl mice, unlike WT mice, are susceptible to ZIKV infection.
Figure 2. The LysMCre+IFNARfl/fl mouse model of ZIKV infection.
WT and LysMCre+IFNARfl/fl C57BL/6 mice at 5 weeks of age were infected with 106 FFU of MR766 or FSS13025. Serum, liver, spleen, and brain were harvested at day 1 and 3 post-infection, and the levels of infectious ZIKV were determined using BHK-21 cell-based FFA. The quantities of infectious (A) MR766 or (B) FSS13025 virus at day 1 (black circles) and day 3 (white squares) post-infection are shown. Four mice were included in each group. (C) Weight and clinical scores of infected WT and LysMCre+IFNARfl/fl mice were monitored and unpaired t test with Welch’s correction was used to compare the two groups at each time point. (D) A representative density plot showing CD44 and CD62L expression and (E) the frequency of CD3+CD8+ T cells and CD44+CD62L− CD8+ T cells from LysMCre+IFNARfl/fl mice infected with 104 FFU of ZIKV or MOCK are shown. Kruskall Wallis test was used first to compare all groups and the Mann-Whitney test was used to compare MOCK and each ZIKV-infected group. All error bars correspond to SEM.
To evaluate whether LysMCre+IFNARfl/fl mice demonstrate a clinical phenotype of ZIKV infection, we compared the clinical scores and the weights of LysMCre+IFNARfl/fl vs. WT mice. Using a clinical criteria scale, we observed that LysMCre+IFNARfl/fl mice developed clinical features up to score 3, corresponding to ruffling of their fur. The infection also induced weight loss in LysMCre+IFNARfl/fl mice between days 4 and 7 post-infection (Figure 2C). However, no signs of paralysis, a dominant phenotype of ZIKV-infected IFNAR−/− mice, and death were observed.
We next explored CD8+ T cell expansion and activation following ZIKV infection of LysMCre+IFNARfl/fl mice. CD44 and CD62L markers differentiated the antigen-experienced CD8+ T cell subset as represented in the gating strategy (Figure 2D) for splenocytes from MOCK and ZIKV-infected (MR766 and FSS13025) mice. ZIKV infection led to an increase in the number of total CD8+ T cells and approximately 5- and 6-fold expansion of CD44+CD62L− CD8+ T cells in mice infected with MR766 and FSS13025, respectively, as compared to uninfected mice (Figure 2E). Thus, LysMCre+IFNARfl/fl mice mount a robust CD8+ T cell response to ZIKV infection.
Identification and validation of ZIKV-derived epitopes recognized by CD8+ T cells in LysMCre+IFNARfl/fl mice
All 244 peptides were tested by IFNγ-ELISPOT assay using CD8+ T cells from ZIKV-infected mice. Fifteen peptides were statistically positive for both MR766 (Figure 3A, top panel) and FSS13025 (Figure 3A, bottom panel). Eight and 7 proteins of the 10 ZIKV proteins are represented among these positive peptides from MR766 and FSS13025, respectively: 40% are from E protein, 13% from prM, NS2A, NS3 or NS5, and 6% from NS2B, NS4A, or NS4B for MR766. FSS13025 epitopes are similarly represented as MR766. All epitopes identified by IFNγ-ELISPOT for both ZIKV strains are indicated in Table 1. Fourteen peptides are recognized only by MR766-primed CD8+ T cells, 3 are specific for FSS13025-primed CD8+ T cells, and twelve are recognized by both MR766- and ZIKV FSS13025-primed CD8+ T cells (Table 1).
Figure 3. Identification of ZIKV epitopes recognized by CD8+ T cells in LysMCre+IFNARfl/fl mice.
Five-week-old LysMCre+IFNARfl/fl were infected retro-orbitally with 104 FFU of ZIKV MR766 or FSS13025. (A) IFNγ-ELISPOT was performed using CD8+ T cells isolated from infected mice. A total of 244 peptides from ZIKV strains predicted to bind H-2Kb and H-2Db with high affinity were screened. Two independent experiments for each ZIKV strain (n=5 mice for each experiment) were performed in triplicate per peptide. The data are expressed as the mean of spot forming cells (SFC) per 106 CD8+ T cells, and error bars are represented as SEM. One-way ANOVA was used to compare the mean of each peptide with the control (DMSO) (P > 0.05). (B) To confirm ZIKV-derived epitopes recognized by CD8+ T cells in LysMCre+IFNARfl/fl mice and IFNγ production via ICS was determined seven days post-infection with positive peptides. The dotted line corresponds to the average amount of IFNγ produced by MOCK mice when stimulated with positive ZIKV-derived peptides (0.19 % of IFNγ+CD8+ T cells).
Table 1. Characteristics of ZIKV-derived peptides.
Peptides from MR766 and FSS13025 ZIKV strains were predicted to bind H-2b class I molecules (Db and Kb). The positions, sequences, and lengths of each of the 29 peptides that induced a positive T cell response, as determined via IFNγ-ELISPOT assay, are shown. The sequence conservation among more than 100 ZIKV strains was obtained using the program BLASTP 2.5.1 on NCBI, and 80% of these strains represent 2015–2016 isolates from Japan, Florida, Singapore, Venezuela, Australia, and Brazil. Y corresponds to highly conserved peptides, sharing 100% (Y(100%)) or 80% (Y(80%)) of sequence identity with the majority of the published strains.
| Sequence | Length | Db | Kb | Protein | Start- position |
End- position |
Conserved | Strains |
|---|---|---|---|---|---|---|---|---|
| AAFTFTKV | 8 | X | E | 600 | 607 | Y (100%) | MR766 | |
| AAGAWYVYV | 9 | X | NS2B | 1489 | 1497 | Y (100%) | MR766 | |
| ISFATTLGV | 9 | X | PrM | 145 | 153 | Y (80%) | MR766 | |
| MSYECPML | 8 | X | X | PrM | 171 | 178 | Y (100%) | MR766 |
| PSVRNGNEI | 9 | X | NS3 | 1866 | 1874 | Y (100%) | MR766 | |
| RAIWYMWL | 8 | X | X | NS5 | 2993 | 3000 | Y (100%) | MR766 |
| RQVMNIVSSWL | 11 | X | NS5 | 2892 | 2902 | Y (100%) | MR766 | |
| SSIAARGYI | 9 | X | NS3 | 1795 | 1803 | Y (100%) | MR766 | |
| SSLVNGVVRL | 10 | X | NS5 | 2839 | 2848 | Y (100%) | MR766 | |
| SSWLWKEL | 8 | X | NS5 | 2899 | 2906 | Y (100%) | MR766 | |
| TGWSNWEEV | 9 | X | NS5 | 3220 | 3228 | Y (100%) | MR766 | |
| TTVSNMAEV | 9 | X | E | 338 | 346 | Y (100%) | MR766 | |
| VMIFLSTAV | 9 | X | X | E | 784 | 792 | Y (80%) | MR766 |
| YSLECDPAVI | 10 | X | NS1 | 969 | 978 | Y (100%) | MR766 | |
| AAFTFTKI | 8 | X | X | E | 600 | 607 | Y (100%) | FSS |
| SSLINGVVRL | 10 | X | NS5 | 2839 | 2848 | Y (80%) | FSS | |
| TLGMNKCYI | 9 | X | PrM | 150 | 158 | Y (100%) | FSS | |
| IMVAVGLL | 8 | X | NS4A | 2257 | 2264 | Y (100%) | MR766/FSS | |
| ATMSYECPM | 9 | X | PrM | 169 | 177 | Y (100%) | MR766/FSS | |
| CAEAPNMKVI | 10 | X | NS5 | 2783 | 2792 | Y (100%) | MR766/FSS | |
| IGVSNRDFV | 9 | X | E | 294 | 302 | Y (100%) | MR766/FSS | |
| MAVDMQTLTPV | 11 | X | E | 635 | 645 | Y (100%) | MR766/FSS | |
| RMAVLGDTA | 9 | X | E | 710 | 718 | Y (100%) | MR766/FSS | |
| RSYCYEASI | 9 | X | E | 347 | 355 | Y (100%) | MR766/FSS | |
| SNRDFVEGM | 9 | X | E | 297 | 305 | Y (100%) | MR766/FSS | |
| SQLTPLTLI | 9 | X | NS4B | 2371 | 2379 | Y (100%) | MR766/FSS | |
| SVKKNLPFVM | 10 | X | NS2A | 1336 | 1345 | Y (100%) | MR766/FSS | |
| VSFIFRAN | 8 | X | X | NS2A | 1237 | 1244 | Y (100%) | MR766/FSS |
| VVIKNGSYV | 9 | X | NS3 | 1656 | 1664 | Y (100%) | MR766/FSS |
To verify the map of the CD8+ T cell response to ZIKV, the computational epitope prediction approach was compared to the overlapping peptide method (screening 15-mer peptides that overlap by 11 amino acids in the E protein from both ZIKV strains). One hundred and twenty-seven peptides corresponding to the complete sequences of E proteins of both MR766 and FSS13025 were tested by IFNγ-ELISPOT. In total, 14 and 15 peptides generated by overlap were positive for MR766 and FSS13025, respectively. Six of the 8 computationally predicted MR766 peptides and 4 of the 5 computationally predicted FSS13025-peptides were identified as positive using the overlapping approach (Supplementary Figure 2).
To validate the epitopes identified via the IFNγ-ELISPOT assay, we performed ICS and quantified CD8+ T cell production of IFNγ, TNFα, and CD107a after stimulation with individual peptides. Among all positive peptides identified by IFNγ-ELISPOT assay, 8 peptides from MR766-infected mice (Figure 3B, top panel) and 4 peptides from FSS13025-infected mice (Figure 3B, bottom panel) induced a high frequency of IFNγ+ cells. For MR766, epitopes were derived from prM (25%), E (37%), NS3 (12%) and NS5 (25%). For FSS13025, 50% of epitopes were from the E protein and 16% were from prM, NS2A, and NS5.
Polyfunctionality of CD8+ T cells after peptide stimulation was evaluated based on the frequency of IFNγ+TNFα+ and CD107a+IFNγ+ (gating strategy, Figure 4B) double-positive cells in MR766-primed (Figure 4A and 4C, top panel) and FSS13025-primed CD8+ T cells (Figure 4A and 4C, bottom panel). The results confirmed that prM169–177, E294–302, E297–305, and NS52783–2792 are the immunodominant epitopes. We extended our investigation by assessing granzyme B expression in CD8+ T cells. The percentages of granzyme B+ CD8+ T cells in infected mice were similar for both ZIKV strains for MR766 and FSS13025, and were 15- and 14-fold higher, respectively, relative to uninfected animals (Figure 4D). To verify cytolytic activity of the ZIKV-specific CD8+ T cells, an in vivo cytotoxicity assay was performed using splenocytes pulsed with three immunodominant peptides (prM169—177; E297–305; NS52783–2792) as targets (Figure 4E). As expected, a high percentage of cytotoxicity was observed in both MR766- and FSS13025-infected mice (Figure 4F). Taken together, these results demonstrate that the epitope-specific CD8+ T cells exhibit a polyfunctional phenotype.
Figure 4. Polyfunctional phenotype of ZIKV epitope-specific CD8+ T cells in LysMCre+IFNARfl/fl mice.
Splenocytes from LysMCre+IFNARfl/fl mice that were infected with 104 FFU of ZIKV strain MR766 (n=4) or FSS13025 (n=5). (A) Frequency of CD8+ T cells producing IFNγ and TNFα upon stimulation with MR766-derived peptides (top panel) and FSS13025-derived peptides (bottom panel) is shown. All experiments were performed twice and error bars are represented in SEM. (B) The gating strategy used to select cells expressing both CD107a and IFNγ in MOCK and infected mice upon stimulation with relevant ZIKV-peptide (E294–302) or with irrelevant ZIKV-peptide (E710–718) is represented. (C) Frequency of CD107a+IFNγ+ CD8+ T cells obtained after stimulation with MR766- or FSS13025-derived peptides is shown. The background obtained in MOCK mice is represented by dotted lines and corresponded to 0.07 % for IFNγ+TNFα+ (panel A) and 2.36 % for CD107a+IFNγ+ double-positive CD8+ T cells (panel C). (D) The percentage of granzyme B produced by CD8+ T cells from mice infected with MR766 (n=8) or FSS13025 ZIKV(n=8) and MOCK (n=4) is represented. (E) A representation of In vivo cytotoxicity of target cells in ZIKV-infected mice is shown. (F) The percentage of killing was obtained in mice infected with ZIKV (n=4) for 7 days or in MOCK (n=4).
Kinetics of the ZIKV-specific CD8+ T cell response in LysMCre+IFNARfl/fl mice
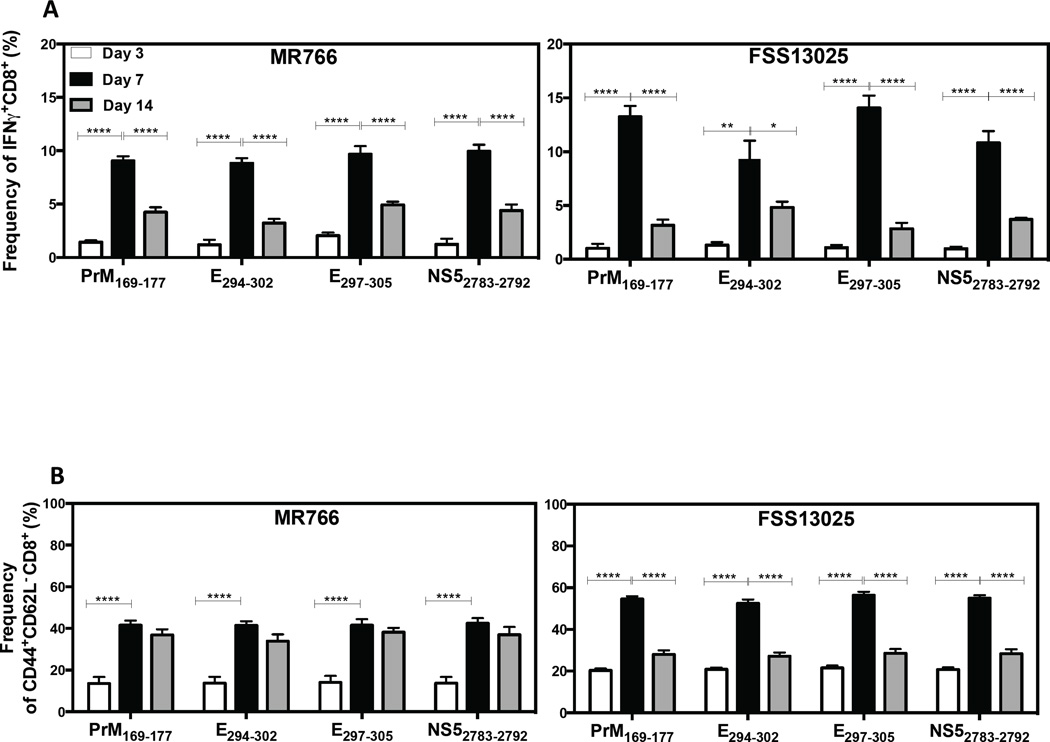
The kinetics of the splenic CD8+ T cell response induced by the immunodominant epitopes at days 3, 7 and 14 post-infection were measured in LysMCre+IFNARfl/fl mice infected with MR766 or FSS13025. The percentage of IFNγ+ CD8+ T cells was higher at day 7 than day 3 or day 14 post-infection for both MR766 and FSS13025-infected mice (Figure 5A). Similarly, the frequency of CD44+CD62L− cells in infected mice was higher at day 7 than day 3 (Figure 5B). These results demonstrate that, among the time points measured, the CD8+ T cell response in ZIKV-infected mice peaks at day 7 post-infection.
Figure 5. Kinetics of the ZIKV-specific CD8+ T cell response in LysMCre+IFNARfl/fl mice.
LysMCre+IFNARfl/fl mice were infected with 104 FFU of ZIKV strain MR766 or FSS13025. Splenocytes were harvested at 3, 7, and 14 days post-infection and stimulated with immunodominant ZIKV-derived peptides to assess cytokine production by ICS. (A) The frequency of IFNγ-producing CD8+ T cells and (B) the frequency of CD44+CD62L− CD8+ T cells at day 3 (white), 7 (black), and 14 (grey) post-infection are shown. The background production of IFNγ obtained in MOCK was subtracted from all values. Two-way ANOVA test was used to compare the time points for each peptide (P > 0.05). The error bars correspond to SEM.
CD8+ T cells control ZIKV infection in LysMCre+IFNARfl/fl mice
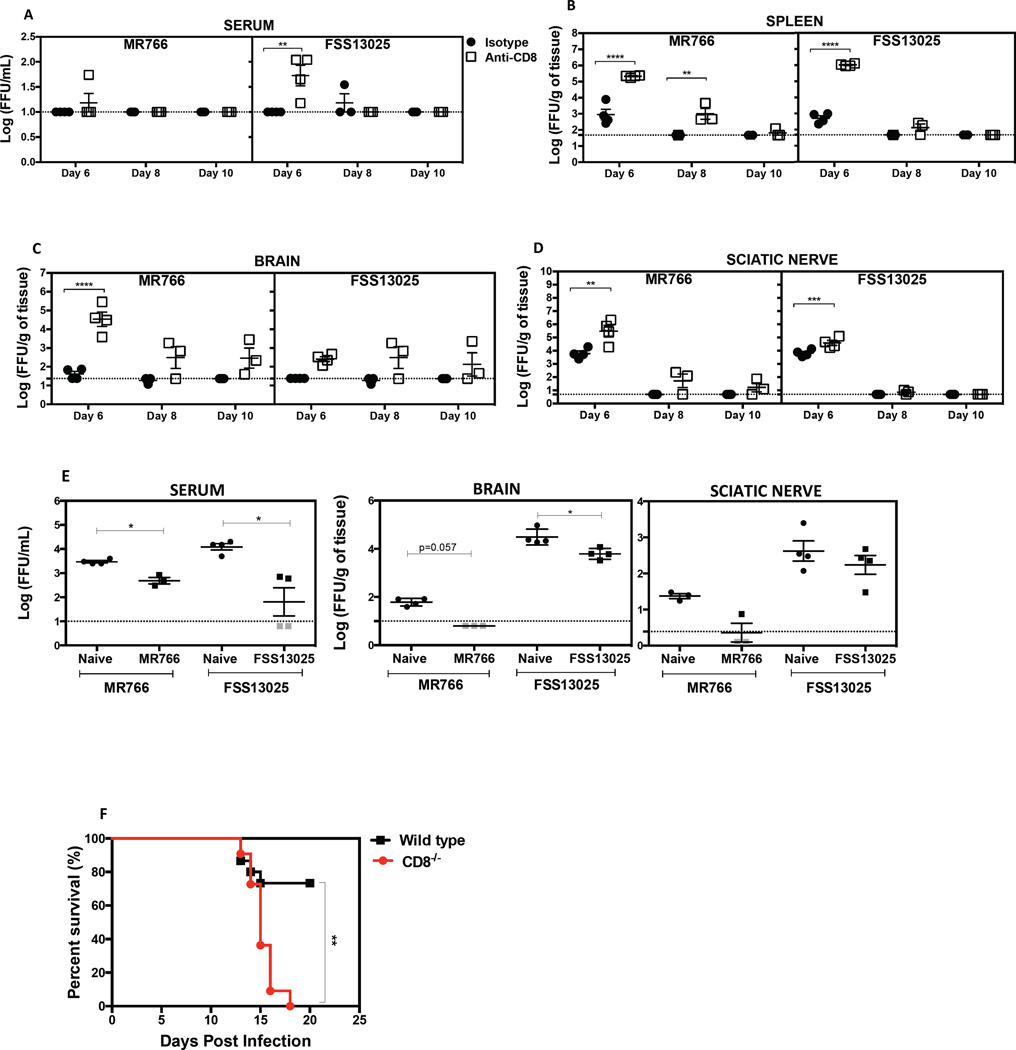
We next explored the role of CD8+ T cells in controlling ZIKV infection by performing antibody-mediated depletion studies. Levels of infectious virus in serum (Figure 6A), spleen (Figure 6B), brain (Figure 6C) and sciatic nerve (Figure 6D) were assessed 6, 8 or 10 days after infection of CD8+ T cell-sufficient and -depleted mice. At day 6 post-infection, CD8+ T cell-depleted mice infected with MR766 or FSS13025 contained higher viral burdens in the serum, spleen, brain, and sciatic relative to the CD8+ T cell-sufficient control mice. At day 8 post-infection, the amount of virus decreased in all tissues, in both control and CD8-depleted groups. At day 10, the level of infectious ZIKV was undetectable in almost all of the tissues (Figures 6A, 6B, and 6D) except the brain (Figure 6C).
Figure 6. Protective role of CD8+ T cells against ZIKV infection in mice.
LysMCre+IFNARfl/fl were treated with depleting anti-CD8 or isotype control antibody on days 3 and 1 before infection with 105 FFU of MR766 or FSS13025. Mice were sacrificed and tissues harvested at 6, 8 and 10 days post-infection. The levels of infectious virus in the (A) serum, (B) spleen, (C) brain, and (D) sciatic nerve were quantified using BHK-21 cell-based FFA. A two-way ANOVA test was used to compare the levels of infectious ZIKV between the isotype and the anti-CD8 antibody-administered groups for all time points and tissues. (E) On day 120 after infection with MR766 or FSS13025, 7.5 × 106 CD8+ T cells were transferred into 5-week-old naive mice one day before challenge with 105 FFU of MR766 or FSS13025. For controls, CD8+ T cells were isolated from naive LysMCre+IFNARfl/fl mice. Infectious ZIKV was quantified. Mann-Whitney test was used to compare naïve CD8+ T cells vs. ZIKV-immune CD8+ T cells. (F) Seven-week-old WT and CD8α−/− were treated with 2 mg of IFNAR-blocking antibody at day -1, and then inoculated subcutaneously with 105 PFU of mouse adapted Dakar 41519 ZIKV strain at day 0. Survival was monitored for 21 days in both groups and reported for WT (n=15, Black square) and CD8−/− (n=11, Red circle) mice. Pooled data from three independent experiments are represented and the log-rank (Mantel-cox) test was used to compare groups.
Next, we adoptively transferred memory CD8+ T cells from LysMCre+IFNARfl/fl donor mice infected with MR766 or FSS13025 for 120 days. ZIKV-immune memory CD8+ T cells were transferred to naïve recipient LysMCre+IFNARfl/fl mice one-day prior to infection with MR766 or FSS13025. Transfer of 7.5 × 106 memory CD8+ T cells resulted in decreased ZIKV burden compared to control T cells from naïve mice (Figure 6E) in the serum and brain (Figure 6E).
During its generation in the 1940s and 1950s, ZIKV MR766 was passaged serially more than 100 times in mouse brains, leading to a neurologically adapted virus (Haddow et al., 2012). To confirm the role of CD8+ T cells during ZIKV infection using a second ZIKV strain of African lineage as well as another loss-of-function model for CD8+ T cells, we performed a survival study using mouse-adapted ZIKV strain Dakar 41519 and Cd8a gene-deficient mice lacking CD8+ T cells. Survival was monitored in IFNAR-blocking antibody-treated WT and congenic CD8+ T cell-deficient (CD8−/−) C57BL/6 mice (Figure 6F). Mice started to die 12 days after infection with mouse-adapted ZIKV Dakar 41519. Eighteen days later, all CD8−/− mice were dead compared to only 25% of WT mice. Thus, a lack of CD8+ cells significantly increased susceptibility to lethal ZIKV infection. Collectively, our results demonstrate a critical role for CD8+ T cells in controlling ZIKV infection and pathogenesis in mice.
DISCUSSION
We conclude that CD8+ T cells play a protective role against ZIKV infection in an animal model with IFN receptor-competent T cells and dendritic cells, and that the specificity of the CD8+ T cell response varies slightly among ZIKV strains. We provide a validated map of the CD8+ T cell response to ZIKV strains MR766 and FSS130125 with identification of 26 and 15 epitopes, respectively. Moreover, all three immunodominant peptides are highly conserved. These maps establish a foundation for investigating CD8+ T cell responses to ZIKV. Our results suggest that an effective ZIKV vaccine should induce a broad CD8+ T cell response.
ZIKV publications through November 2016 have not described any data on the T cell response to ZIKV in humans or animal models. Currently DENV mouse models provide the largest body of information regarding CD8+ T cell responses to systemic Aedes-transmitted flavivirus infection. Similar to our present results with ZIKV, a protective role for CD8+ T cells against DENV was established as increased viral loads were observed following CD8+ T cell depletion (Yauch et al., 2009). In addition, adoptive transfer of DENV-primed CD8+ T cells (Zellweger et al., 2014) and effective epitope vaccination studies (Yauch et al., 2009) provided further indication of CD8+ T cells’ protective role against DENV.
Our data showed broad CD8+ T cell responses to ZIKV MR766 and FSS13025 that target all viral proteins with the exception of NS1 and NS2B in the FSS13025 response. In H-2b mice, E protein appeared to be the main target of the anti-ZIKV CD8+ T cell response, whereas for DENV dominant epitopes are within NS3, NS4B, and NS5 (Weiskopf et al., 2013; Yauch et al., 2009). When grouped by protein, epitope immunodominance between the two ZIKV strains was similar for the prM, E, and NS5 epitopes. However, a stronger response was seen for MR766 NS31866–1874. Overall, the H-2b CD8+ T cell response to MR766 was broader than to FSS13025, especially for NS1, NS3, and NS5 epitopes.
The contribution of CD8+ T cells to protection vs. pathogenesis in ZIKV infection, and whether cross-reactive antibodies or T cells can worsen the course of ZIKV infection following infection with a similar flavivirus through antibody-dependent enhancement or original T cell antigenic sin, respectively, remain to be determined (Lazear and Diamond, 2016). However, evidence of these phenomena (Halstead, 2007; Mongkolsapaya et al., 2003) from cases of severe DENV would indicate that vaccine developers need to consider the effects of ZIKV vaccine if recipients subsequently become infected with DENV. Therefore the use of epitopes to design a subunit vaccine may be a good alternative for ZIKV.
Our findings lay the groundwork for investigating the function of CD8+ T cells in ZIKV infection of immunologically specialized sites. Unlike dengue disease in which systemic infection dominates the clinical course (Mongkolsapaya et al., 2006), it is the localized events preceding and following systemic infection that are the most threatening components of ZIKV’s clinical picture. Documentation of mucosal transmission by vaginal and anal intercourse leading to systemic infection is growing (D'Ortenzio et al., 2016; Deckard et al., 2016; Foy et al., 2011; Hills et al., 2016; Musso et al., 2015; Venturi et al., 2016). Once maternal systemic infection has been established, transplacental infection and transmission allows for infection of the fetal brain and devastating consequences including microcephaly (Brasil et al., 2016; Lazear and Diamond, 2016; Malone et al., 2016; Mlakar et al., 2016; Oliveira Melo et al., 2016; Tetro, 2016; Ventura et al., 2016). Post-systemic infection entry of the virus into semen-producing tissues (Atkinson et al., 2016; Govero et al., 2016) allows the virus to be transmitted without its mosquito vector. Finally, evidence is also mounting that autoimmune disease such as GBS can follow systemic infection with ZIKV (Deckard et al., 2016; Lazear and Diamond, 2016; Malone et al., 2016; Oehler et al., 2014).
ZIKV’s most devastating clinical effects result from infection of the fetal brain, and CD8+ T cell-dependent clearance of other neurotropic flaviviruses is well documented (Shrestha and Diamond, 2004). ZIKV burden observed in the brains of both CD8+ T cell-sufficient and -depleted LysMCre+IFNARfl/fl mice is consistent with published evidence of ZIKV’s neurotropism in mice (Cugola et al., 2016; Dowall et al., 2016; Lazear et al., 2016; Li et al., 2016a; Li et al., 2016b; Miner et al., 2016; Rossi et al., 2016). In the latter mice, disproportionately increased levels of ZIKV MR766 in the brain seen at day 6 post-infection may reflect the strain’s passage history through mouse brains (Dick, 1952). This observation in the brains of mice infected with MR766 relative to FSS13025 highlights one of the differences between these two strains, albeit this difference was not observed at earlier time points (days 1 and 3 post-infection).
The susceptibility of LysMCre+IFNARfl/fl mice to ZIKV indicates that loss of type I IFN response in myeloid cells is sufficient to permit robust ZIKV infection. This finding is consistent with reported permissiveness of monocytes and macrophages to replication of other flaviviruses (Mangada et al., 2002; Prestwood et al., 2012a; Shrestha et al., 2008; Yang et al., 2014). Transplacental ZIKV transmission was recently reported in SJL mice, which also have an intact IFN response, but no mention of the specific cellular tropism was made (Cugola et al., 2016). The exact effects of the myeloid cell type I IFN response on the anti-ZIKV CD8+ T cell response remain unknown. Similar to Zika fever in adult humans (Lazear and Diamond, 2016; Malone et al., 2016), LysMCre+IFNARfl/fl mice become transiently ill (ruffled coat, hunched posture, weight loss) and recover from clinical signs at day 6–7 after infection, which corresponds to peak CD8+ T cell IFNγ and CD107a expression. Based on CD8+ T cell expansion, polyfunctional phenotype of ZIKV epitope-specific CD8+ T cells, and CD8+ T cell-mediated viral clearance, we conclude that myeloid type I IFN response is not necessary for priming an efficient CD8+ T cell response in these mice. This conclusion is similar to studies of DENV (Yauch et al., 2009), vaccinia virus, vesicular stomatitis virus (Thompson et al., 2006), and Sendai virus (Lopez et al., 2006) in IFNAR-deficient mice. Further characterization of the LysMCre+IFNARfl/fl mouse model should provide a platform for studying ZIKV-specific T cell responses, and for testing vaccine and antiviral candidates.
EXPERIMENTAL PROCEDURES
Viral strains and mice
ZIKV strains MR766 and FSS13025 were obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). MR766, African lineage was isolated from a sentinel monkey rhesus (766) in east Africa (Dick, 1952). Since this isolation, the MR 766 isolate has been passaged over 100 times in mice using intracerebral inoculations (Dick, 1952). ZIKV FSS13025 was isolated in 2010 from a Cambodian pediatric case (Heang et al., 2012) and has been passaged a low number of times. We cultured MR766 and FSS13025 using C6/36 Aedes albopictus mosquito cells as described previously (Prestwood et al., 2008). Virus was harvested from cell supernatants 7–10 days after infection, followed by clarification via centrifugation, and concentration via ultracentrifugation as previously described (Prestwood et al., 2012a). Virus was titrated using Baby hamster kidney (BHK)-21 cell-based focus-forming assay (FFA). ZIKV strain Dakar 41519, isolated in Senegal in 1984, was also obtained from WRCEVA, passaged four times in RAG−/− mice and amplified once in Vero cells (African green Monkey Kidney Epithelial Cells) as described (Govero et al., 2016; Sapparapu et al., 2016). Next generation sequencing of ZIKV stocks confirmed the sequence of each strain and the absence of adventitious pathogens.
Wild type mice were purchased from the Jackson laboratories, and LysMCre+IFNARfl/fl and CD8α−/− C57BL/6 mice were bred at La Jolla Institute for Allergy & Immunology and Washington University School of Medicine Animal Facilities. WT mice were treated with 1 or 2 mg of mouse anti-IFNAR (MAR1-5A3) depending on the experiment or isotype control (MOPC-21) monoclonal antibody one day prior to infection. All experiments were performed following the institutional Animal Care and Use Committee-approved animal protocols. Both male and female mice between 5–7 weeks of age were used in this study and all in vivo infections were performed either retroorbital or subcutaneous inoculations with 200 µl of ZIKV in 10% FBS/PBS buffer containing 104, 105, or 106 Focus Forming Units (FFU) of virus. In all experiments, mice receiving 10 % FBS/PBS buffer, are designated as MOCK. For survival study, mice were infected with 50 µl of Dakar 41519 ZIKV strain diluted in PBS.
To assess the clinical features, mice were checked each day and assigned a score between 1 and 7 as previously described (Tang et al., 2016). Weights were recorded, reported and compared to the initial weight obtained on the day of infection.
Titration of virus by FFA
BHK-21 cells were plated at 2 × 105 cells per well in a 24-well plate and incubated at 37°C, 5% CO2 overnight. Following mouse perfusion with PBS, organs were harvested in 1 ml of MEM-α-medium (Invitrogen) in pre-weighed tubes containing steel beads, followed by homogenization and then centrifugation at 2000g for 5 minutes. The clarified supernatant was used to infect BHK cells following serial dilution, and cells were infected for 1 hour with gentle shaking every 15 minutes. After infection, wells were overlaid with carboxymethyl cellulose (CMC) (Sigma). Two days after infection, cells were fixed with 4% formalin (Fisher Chemicals), permeabilized with 1% Triton X (Sigma), and blocked with 10% FBS-PBS. Viral antigen was detected using 4G2, a pan-flavivirus anti-envelope (E) antibody, following by a secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma). Foci were revealed after incubation with True Blue substrate (KPL) and were counted manually.
Peptide prediction approaches
All known ZIKV polyprotein sequences for African and Asian lineages were obtained from the NCBI protein database in January 2016. MHC class I-peptide binding affinity predictions were performed at the Immune Epitope Database (IEDB) Tools website using ‘IEDB-recommended’ method selection, as previously described (Kim et al., 2012). Predicted binding affinities for all non-redundant 8-11mer peptides that bound H2-Kb and H2-Db were obtained. For each allele, the lists of peptides obtained above were sorted by increasing consensus percentile rank and restricted to the top 1%.
The E protein from ZIKV strains MR766 and FSS13025 was selected to identify epitopes using the overlapping methods (Supplementary Figure 2). 15-mer peptides that overlapped by 11 amino acids were designed from the E protein sequence and synthesized.
Peptide synthesis
All peptides were synthesized by Synthetic Biomolecules, San Diego, CA. All 9-, 10-, 11- and 15-mer peptides for ELISPOT were synthesized as crude material on a 1-mg scale and mass spectral analysis of each peptide was performed to validate the synthesis. Peptides for flow cytometric analyses were synthesized and purified by reverse-phase HPLC to ≥ 95% purity. Peptides were dissolved in DMSO and aliquoted.
Ex vivo gamma interferon (IFNγ) ELISPOT
CD8+ T cells were isolated by magnetic bead positive selection (Miltenyi Biotec, Germany). A total of 2 × 105 CD8+ T cells were stimulated with 1 × 105 LPS-blast cells as antigen presenting cells (APCs) and 10 µg of individual ZIKV-derived peptide in 96-well flat-bottom plates (Immobilon-P; Millipore, MA) coated with anti-mouse IFNγ monoclonal antibody (mAb) (clone AN18; Mabtech, Sweden) in triplicate. IFNγ-ELISPOT was performed as previously detailed (Elong Ngono et al., 2016). Positive peptides were those with a number of spot-forming cells (SFC) per 106 CD8+ T cells ≥ 20 and a stimulation index ≥ 2 based on the negative control (DMSO).
Flow cytometric analyses
For ICS, splenocytes were counted after red blood cell lysis and resuspended in 10% FBS/RPMI medium at 40 × 106 cells per ml. Splenocytes (2 × 106) were plated and stimulated with 1 µg of individual peptide as previously detailed (Elong Ngono et al., 2016). Positive (PMA-Ionomycin) and negative (No stimulation) controls were added for all experiments. Cells were labeled with anti-CD3 (Clone 145-2C11), anti-CD8 (clone 53–67), anti-CD44 (clone IM7), and anti-CD62L (clone Mel-14). Cells then were fixed and permeabilized, followed by staining with anti-granzyme B (clone NGZB), anti–IFNγ (clone XMG 1.2) and anti-TNFα (clone MP6-XT22). Samples were read on an LSR II (BD Biosciences) and were analyzed using FlowJo software X 10.0.7 (Tree Star, Ashland, OR).
In vivo cytotoxicity assay
LysMCre+IFNARfl/fl and WT mice were infected with 104 FFU of MR766 or FSS13025. Seven days post-infection, splenocytes were harvested from naïve donor mice, followed by stimulation with a pool of H-2b-restricted ZIKV-peptides (PrM169–177, E297–305, NS52783–2792) referred to as “Target Cells” or with DMSO for 3 h at 37°C. The cells were washed and labeled with CSFE (Invitrogen) in PBS/0.1% BSA for 10 min at 37°C. Target cells were labeled with 1 µM CSFE (High) or 100 nM CSFE (Low) for unstimulated cells. After washing, 107 of labeled cells (5 × 106 of each population) were injected intravenously into MOCK and infected recipients. Splenocytes from recipients were harvested 4 (Wild type) or 12 h later (LysMCre+IFNARfl/fl) and analyzed by flow cytometry. The percentage of killing is calculated as followed: 100 - (% ZIKV-peptide stimulated in infected mice / % DMSO-stimulated in infected mice) / (% ZIKV-peptide stimulated in naïve mice / % DMSO-stimulated in naïve mice) × 100).
Depletion and adoptive transfer of CD8+ T cells
All antibodies for depletion studies were purchased from BioXCell. Mice were injected intraperitoneally (i.p.) with CD8 cell-depleting (clone YTS 169.4) or rat IgG2 isotype control (clone LTF-2) antibodies on days 3 and 1 prior to infection with 105 FFU of ZIKV MR766 or ZIKV FSS13025. Organs were harvested at day 6, 8 or 10 after infection and levels of infectious virus were determined using BHK-21 cell-based FFA.
ZIKV-immune CD8+ T cells were isolated from LysMCre+IFNARfl/fl mice on day 120 after infection with 105 FFU of MR766 or FSS13025 using magnetic positive CD8+ T cells selection kit (Miltenyi Biotech, CD8a Ly-2). 7.5 × 106 CD8+ T cells were transferred into 5 week-old naïve mice, and recipient mice were challenged with either MR766 or FSS13025 one day after cell transfer. Viral titers in tissues were measured using BHK-21 cell-based FFA three days post-challenge.
Statistical analyses
All data were analyzed with Prism software version 5.0 (GraphPad Software, Inc., San Diego, CA) and expressed as mean ± SEM. Statistical significance was determined using the non-parametric Mann-Whitney test to compare two groups and the Wilcoxon test to compare two parameters from the same group. Two-way ANOVA or the Kruskal-Wallis test was used to compare more than 2 groups. P < 0.05 was considered as significant.
Supplementary Material
Highlights.
LysMCre+IFNARfl/fl mice developed as a model to investigate T cell responses to ZIKV
ZIKV infection elicits CD8+ T cells targeting viral PrM, E and NS5 proteins in mice
ZIKV-immune CD8+ T cell transfer reduced while depletion increased viral burdens in mice
CD8−/− mice displayed higher mortality to ZIKV-infection
Acknowledgments
We thank Jason Greenbaum and Sumetha Kannan for bioinformatics analyses, and Matthew Young and Anila Mamidi for technical assistance with focus-forming assays and breeding/maintaining mice. This research was funded by NIAID/NIH grants (R01 AI116813 (S.S), R01 AI073755 and R01 AI104972 (M.S.D) and the La Jolla Institute for Allergy & Immunology institutional support.
COMPETING INTERESTS
M.S.D. is a consultant for Inbios and Visterra, on the Scientific Advisory Boards of Moderna and OraGene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.E.N., M.S.D., and S.S. designed the studies. A.E.N. designed, performed and analyzed experiments. E.A.V., W.W.T., N.S., Y.J., and M.J.G. performed experiments. A.E.N., K.K, and S.S. interpreted the data and wrote the manuscript. M.S.D and S.S. edited the manuscript.
REFERENCES
- Atkinson B, Hearn P, Afrough B, Lumley S, Carter D, Aarons EJ, Simpson AJ, Brooks TJ, Hewson R. Detection of Zika Virus in Semen. Emerg Infect Dis. 2016;22:940. doi: 10.3201/eid2205.160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Jr, Raja Gabaglia C, Damasceno L, Wakimoto M, Ribeiro Nogueira RM, Carvalho de Sequeira P, Machado Siqueira A, Abreu de Carvalho LM, Cotrim da Cunha D, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro - Preliminary Report. N Engl J Med. 2016 doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016 doi: 10.1038/nature18296. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ortenzio E, Matheron S, de Lamballerie X, Hubert B, Piorkowski G, Maquart M, Descamps D, Damond F, Yazdanpanah Y, Leparc-Goffart I. Evidence of Sexual Transmission of Zika Virus. N Engl J Med. 2016 doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- Deckard DT, Chung WM, Brooks JT, Smith JC, Woldai S, Hennessey M, Kwit N, Mead P. Male-to-Male Sexual Transmission of Zika Virus - Texas, January 2016. MMWR Morb Mortal Wkly Rep. 2016;65:372–374. doi: 10.15585/mmwr.mm6514a3. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. The Journal of experimental medicine. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46:521–534. doi: 10.1016/0035-9203(52)90043-6. [DOI] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. A Susceptible Mouse Model for Zika Virus Infection. PLoS Negl Trop Dis. 2016;10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, Weiskopf D, Sidney J, Sette A, Shresta S. Protective Role of Cross-Reactive CD8 T Cells Against Dengue Virus Infection. EBioMedicine. 2016;13:284–293. doi: 10.1016/j.ebiom.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, et al. Zika virus infection damages the testes in mice. Nature. 2016 doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, Vazques S, Delgado I, Halstead SB. Epidemiologic studies on Dengue in Santiago de Cuba, 1997. American journal of epidemiology. 2000;152:793–799. doi: 10.1093/aje/152.9.793. discussion 804. [DOI] [PubMed] [Google Scholar]
- Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Dengue. Lancet (London, England) 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Heang V, Yasuda CY, Sovann L, Haddow AD, Travassos da Rosa AP, Tesh RB, Kasper MR. Zika virus infection, Cambodia, 2010. Emerg Infect Dis. 2012;18:349–351. doi: 10.3201/eid1802.111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills SL, Russell K, Hennessey M, Williams C, Oster AM, Fischer M, Mead P. Transmission of Zika Virus Through Sexual Contact with Travelers to Areas of Ongoing Transmission - Continental United States, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, Zhao Z. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol. 2007;81:10081–10091. doi: 10.1128/JVI.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioos S, Mallet HP, Leparc Goffart I, Gauthier V, Cardoso T, Herida M. Current Zika virus epidemiology and recent epidemics. Medecine et maladies infectieuses. 2014;44:302–307. doi: 10.1016/j.medmal.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, Lundegaard C, Sette A, Lund O, Bourne PE, et al. Immune epitope database analysis resource. Nucleic acids research. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor Ldel C. Phylogeny of Zika Virus in Western Hemisphere, 2015. Emerg Infect Dis. 2016;22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Diamond MS. Zika Virus: New Clinical Syndromes and its Emergence in the Western Hemisphere. J Virol. 2016 doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear Helen M, Govero J, Smith Amber M, Platt Derek J, Fernandez E, Miner Jonathan J, Diamond Michael S. A Mouse Model of Zika Virus Pathogenesis. Cell Host & Microbe. 2016 doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell stem cell. 2016a;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Tang W, Chai G, JA R-N, Sheets N, AV T, S S, JG G. Zika virus infection in adult brain shows tropism for neural progenitors and alters proliferation. Cell stem cell. 2016b doi: 10.1016/j.stem.2016.08.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CB, Yount JS, Hermesh T, Moran TM. Sendai virus infection induces efficient adaptive immunity independently of type I interferons. J Virol. 2006;80:4538–4545. doi: 10.1128/JVI.80.9.4538-4545.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RW, Homan J, Callahan MV, Glasspool-Malone J, Damodaran L, Schneider Ade B, Zimler R, Talton J, Cobb RR, Ruzic I, et al. Zika Virus: Medical Countermeasure Development Challenges. PLoS Negl Trop Dis. 2016;10:e0004530. doi: 10.1371/journal.pntd.0004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangada MM, Endy TP, Nisalak A, Chunsuttiwat S, Vaughn DW, Libraty DH, Green S, Ennis FA, Rothman AL. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J Infect Dis. 2002;185:1697–1703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell. 2016;165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, Khemnu N, Tangthawornchaikul N, Chotiyarnwong P, Sae-Jang K, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika virus infection complicated by Guillain-Barre syndrome--case report, French Polynesia, December 2013. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2014;19 doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Oliveira Melo AS, Malinger G, Ximenes R, Szejnfeld PO, Alves Sampaio S, Bispo de Filippis AM. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg? Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2016;47:6–7. doi: 10.1002/uog.15831. [DOI] [PubMed] [Google Scholar]
- Pinto AK, Brien JD, Lam CY, Johnson S, Chiang C, Hiscott J, Sarathy VV, Barrett AD, Shresta S, Diamond MS. Defining New Therapeutics Using a More Immunocompetent Mouse Model of Antibody-Enhanced Dengue Virus Infection. mBio. 2015;6:e01316–e01315. doi: 10.1128/mBio.01316-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwood TR, May MM, Plummer EM, Morar MM, Yauch LE, Shresta S. Trafficking and replication patterns reveal splenic macrophages as major targets of dengue virus in mice. J Virol. 2012a;86:12138–12147. doi: 10.1128/JVI.00375-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, Shresta S. Gamma interferon (IFN-gamma) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-alpha/beta receptor-deficient mice. J Virol. 2012b;86:12561–12570. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol. 2008;82:8411–8421. doi: 10.1128/JVI.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. Characterization of a Novel Murine Model to Study Zika Virus. Am J Trop Med Hyg. 2016 doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. American journal of epidemiology. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- Sapparapu G, Fernandez E, Kose N, Cao B, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016 doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. 2015;15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Diamond MS. Role of CD8+ T cells in control of West Nile virus infection. J Virol. 2004;78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Zhang B, Purtha WE, Klein RS, Diamond MS. Tumor necrosis factor alpha protects against lethal West Nile virus infection by promoting trafficking of mononuclear leukocytes into the central nervous system. J Virol. 2008;82:8956–8964. doi: 10.1128/JVI.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Grewal R, Shresta S. Influence of antibodies and T cells on dengue disease outcome: insights from interferon receptor-deficient mouse models. Current opinion in virology. 2015;13:61–66. doi: 10.1016/j.coviro.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Tang WW, Young MP, Mamidi A, Regla-Nava JA, Kim K, Shresta S. A mouse model of Zika virus sexual transmission and vaginal viral replication. Cell Reports. 2016 doi: 10.1016/j.celrep.2016.11.070. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetro JA. Zika and microcephaly: Causation, correlation, or coincidence? Microbes Infect. 2016 doi: 10.1016/j.micinf.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Thompson LJ, Kolumam GA, Thomas S, Murali-Krishna K. Innate inflammatory signals induced by various pathogens differentially dictate the IFN-I dependence of CD8 T cells for clonal expansion and memory formation. J Immunol. 2006;177:1746–1754. doi: 10.4049/jimmunol.177.3.1746. [DOI] [PubMed] [Google Scholar]
- Ventura CV, Maia M, Bravo-Filho V, Gois AL, Belfort R., Jr Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet (London, England) 2016 doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- Venturi G, Zammarchi L, Fortuna C, Remoli ME, Benedetti E, Fiorentini C, Trotta M, Rizzo C, Mantella A, Rezza G, et al. An autochthonous case of Zika due to possible sexual transmission, Florence, Italy, 2014. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.8.30148. [DOI] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci U S A. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Sette A. T-cell immunity to infection with dengue virus in humans. Front Immunol. 2014;5:93. doi: 10.3389/fimmu.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Rainone A, Shi XQ, Fournier S, Zhang J. A new animal model of spontaneous autoimmune peripheral polyneuropathy: implications for Guillain-Barre syndrome. Acta neuropathologica communications. 2014;2:5. doi: 10.1186/2051-5960-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, Shresta S. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. 2010;185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammarchi L, Stella G, Mantella A, Bartolozzi D, Tappe D, Gunther S, Oestereich L, Cadar D, Munoz-Fontela C, Bartoloni A, et al. Zika virus infections imported to Italy: clinical, immunological and virological findings, and public health implications. J Clin Virol. 2015;63:32–35. doi: 10.1016/j.jcv.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Zellweger RM, Eddy WE, Tang WW, Miller R, Shresta S. CD8+ T cells prevent antigen-induced antibody-dependent enhancement of dengue disease in mice. J Immunol. 2014;193:4117–4124. doi: 10.4049/jimmunol.1401597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Miller R, Eddy WE, White LJ, Johnston RE, Shresta S. Role of humoral versus cellular responses induced by a protective dengue vaccine candidate. PLoS Pathog. 2013;9:e1003723. doi: 10.1371/journal.ppat.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Shresta S. Mouse models to study dengue virus immunology and pathogenesis. Front Immunol. 2014;5:151. doi: 10.3389/fimmu.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, Shresta S. CD8+ T Cells Can Mediate Short-Term Protection against Heterotypic Dengue Virus Reinfection in Mice. J Virol. 2015;89:6494–6505. doi: 10.1128/JVI.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.