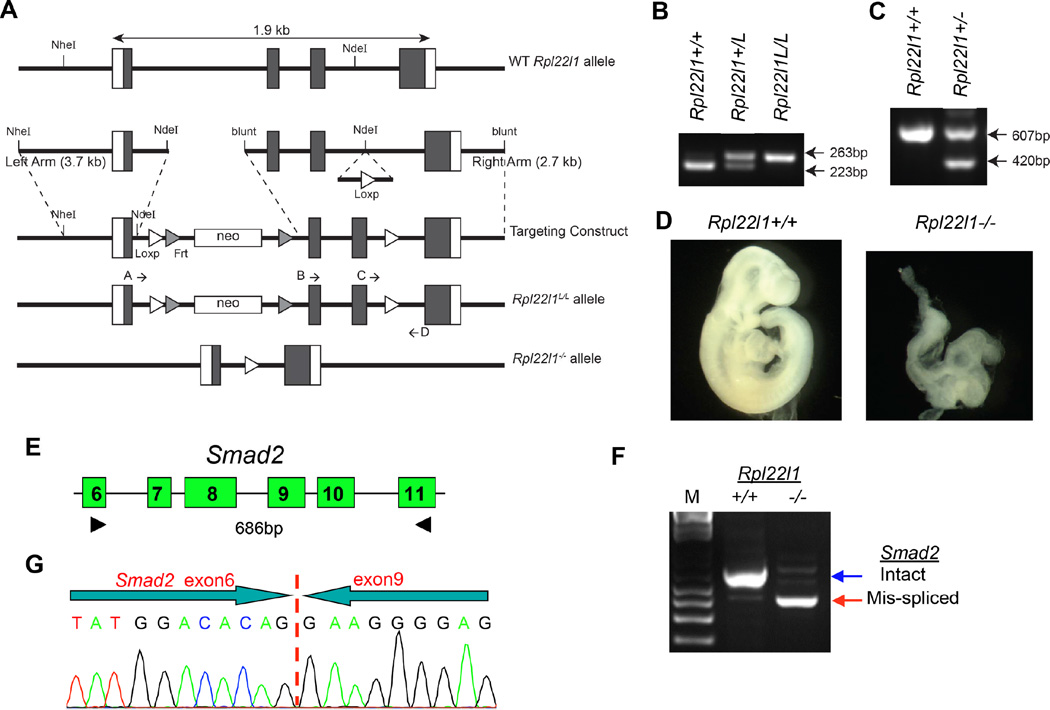
Figure 5. Effect of Like1-deficiency on Smad2 splicing during murine gastrulation.
(A) Molecular strategy for targeted deletion of Rpl22l1. A Loxp-FRT-neo resistance-FRT cassette was inserted 3’ to the first exon of Rpl22l1 and a second LoxP site was inserted 3’ to the third exon. F1 heterozygous offspring were bred to Mox2-cre mice to delete the 2nd and 3rd exons of Rpl22l1, disrupting expression at the genomic locus. (B,C) Strategy to genotype Rpl22l1+/L or Rpl22l1+/− mice. To genotype mice with LoxP sites flanking exons 2 and 3 of Rpl22l1, primers C and D were used to amplify a 223 bp or 263 bp product for wildtype and Rpl22L1-LoxP, respectively. Deletion of Rpl22l1 is genotyped with primers B and D, which amplify a 607 bp product for the WT allele and A and D, which amplify a 420 bp product for the mutant allele after cre recombination. (D) Representative Rpl22l1−/− embryo compared to Rpl22l1+/+ littermate control at 9.5 dpc. (E–G) Effect of Like1-deficiency on splicing of Smad2 pre-mRNA during murine gastrulation. Embryos derived from timed matings of Rpl22l1+/− mice were isolated at 6.5 dpc (mid gastrulation), genotyped as above, and analyzed by RT-PCR using the indicated primers to identify alterations in Smad2 splicing (E,F). Sequencing of the mis-spliced Smad2 species found in Rpl22l1−/− embryos revealed that it represented a species in which exon 6 was fused directly to exon 9, eliminating exons 7 and 8 (G). All results are representative of at least 3 experiments performed.