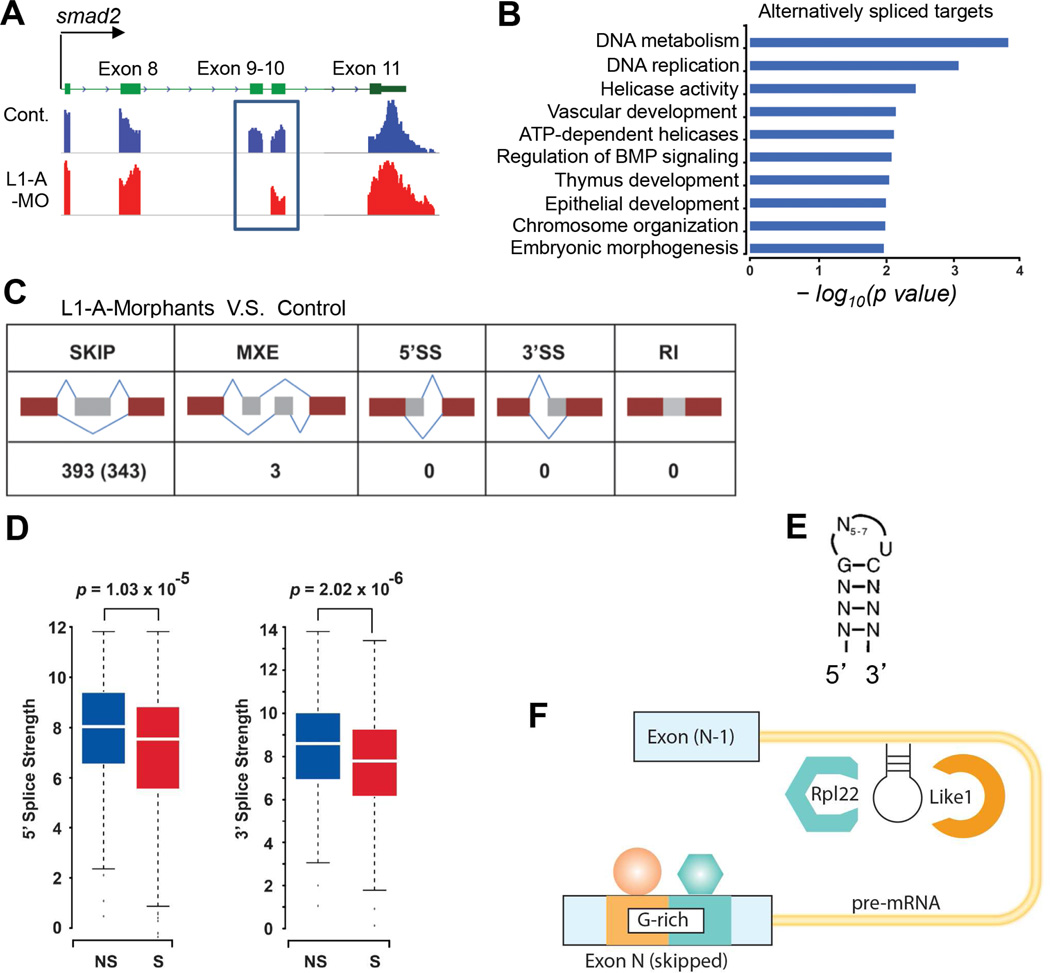
Figure 6. Common features of mRNA targets mis-spliced in Like1 morphants.
(A) smad2 exon9 skipping detected by RNA-Seq. Alignment of RNA-seq reads to the genome reveals exclusion of smad2 exon9 in 10hpf Like1 morphants (blue box). (B) Gene ontology analysis of transcripts affected by Like1 knockdown from RNA-seq analysis. Significant gene ontology terms (p < 0.05) are depicted as a bar graph with the p values represented as –log10 on the X-axis. (C) Schematic illustrating the type of alternative splicing identified by RNA-Seq in 10hpf Like1 morphants. (D) Calculation of the strength of 5´ and 3´ splice sites of skipped (S) versus non-skipped (NS) exons in pre-mRNAs targeted in Like1 morphants. (E) Schematic of the consensus hairpin bound by Rpl22/Like1. (F) Schematic model of the common features of pre-mRNA targets affected by Like1 knockdown. Targets contained both a consensus Rpl22/Like1 binding motif in the preceding intron and a G-rich motif in the skipped exon. RNA-Seq analysis was performed on at least 3 independent biological replicates per condition. See also Figures S5 and S6