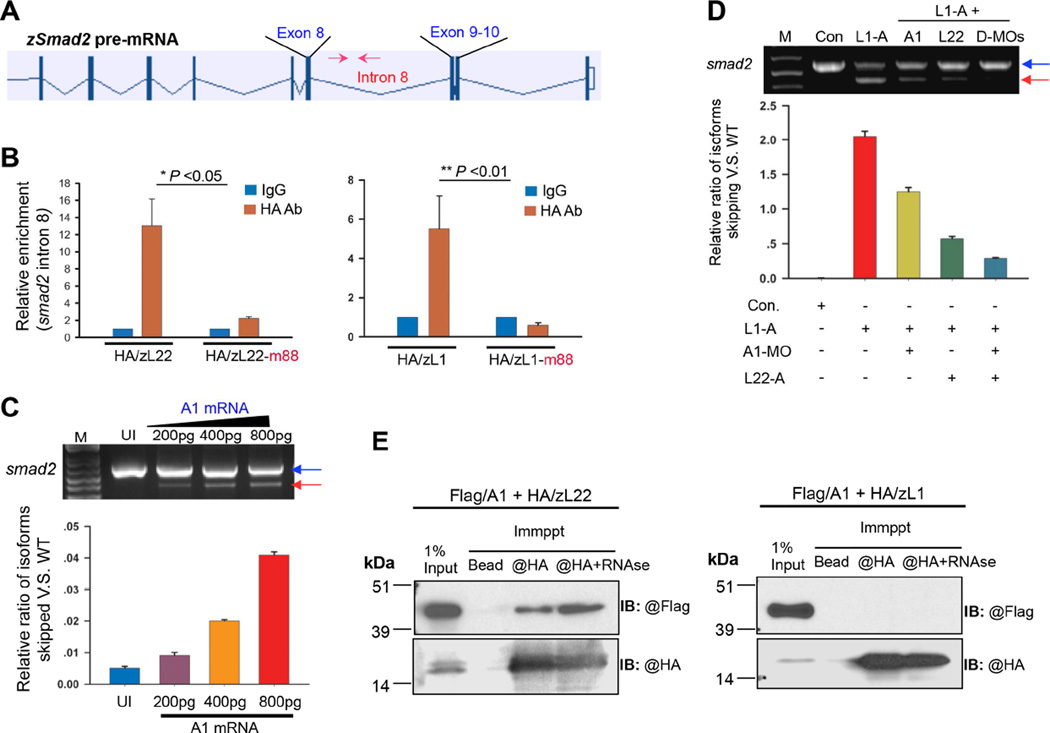
Figure 7.
Factors through which Rpl22 and Like1 regulate smad2 alternative splicing. (A) Position of the real-time primers (red arrows) flanking the consensus Rpl22/Like1 binding site in intron 8 of smad2 pre-mRNA. (B) RNA-CLIP analysis of Rpl22/Like1 binding to smad2 pre-mRNA. Embryos injected with mRNA encoding HA tagged Rpl22, Like1, or their RNA-binding mutants (m88) were harvested at 10hpf. After light crosslinking, detergent nuclear extracts were immunoprecipitated using anti-HA antibody, and the co-precipitated RNA quantified by RT-PCR. Triplicate measurements are depicted graphically as mean ± S.D. p-values are indicated. (C) Overexpression of hnRNP-A1 (A1) mRNA induces smad2 mis-splicing. Embryos were injected with differing amounts of A1 mRNA, following which the effect on smad2 mis-splicing was determined by RT-PCR at 10hpf. The relative ratio of exon 9 skipped mRNA to intact smad2 mRNA was quantified in triplicate and depicted graphically as the mean ± S.D. (D) Genetic interaction of Rpl22 and A1. The ratio of exon9 skipped to intact smad2 was quantified in Like1 morphants in which A1 and/or Rpl22 was knocked down. The mean ± S.D of triplicate measurements was depicted graphically as in (C). (E) Physical association between Rpl22, Like1 and A1. Anti-HA immunoprecipitation (IP) and anti-Flag immunoblots (IB) were performed on detergent extracts of embryos injected with 100pg of mRNA encoding Flag-A1 and either HA-Rpl22 or HA-Like1, either before or after treatment with RNAse. All results are representative of at least 3 experiments performed. See also Figure S7.