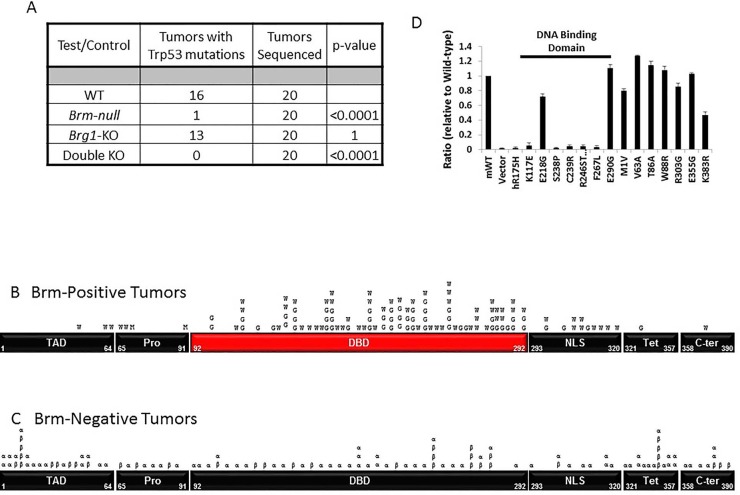
Figure 1. Figure 1A shows the results from sequencing 20 adenocarcinomas from each of the four different mouse phenotypes.
The tumors from Brm-positive mice (wild type and Brg1 knockdown phenotypes) harbored 16/20 and 13/20 Trp53 mutations, respectively, while tumors from Brm-negative mice (Brm-null or double knockout phenotypes) harbored 1/20 and 0/20 Trp53 mutations, respectively. Figures 1B and 1C show the distribution of Trp53 mutations along the Trp53 cDNA from Brm-positive tumors and Brm-negative tumors. The majority of mutations in the Brm-positive tumors are distributed within the Trp53 “hot spot” or DNA binding domain (DBD), while the p53 mutations from the Brm-negative tumors are distributed along the Trp53 cDNA: W=wild type, G=Brg1-KO, α=Brm-null and β=double knockout. Figure 1D fourteen different Trp53 cDNA clones found in both tumors from wild type and Brm-null phenotypes were introduced into the TRP53-negative cell line along with a luciferase TP53 reporter construct. The Trp53 mutations from the DNA binding domain lacked Trp53 transcription activity as measured by this TP53 luciferase reporter as compared to those Trp53 mutations derived from the other Trp53 domains.